Figure 7. A conserved homology search mechanism.
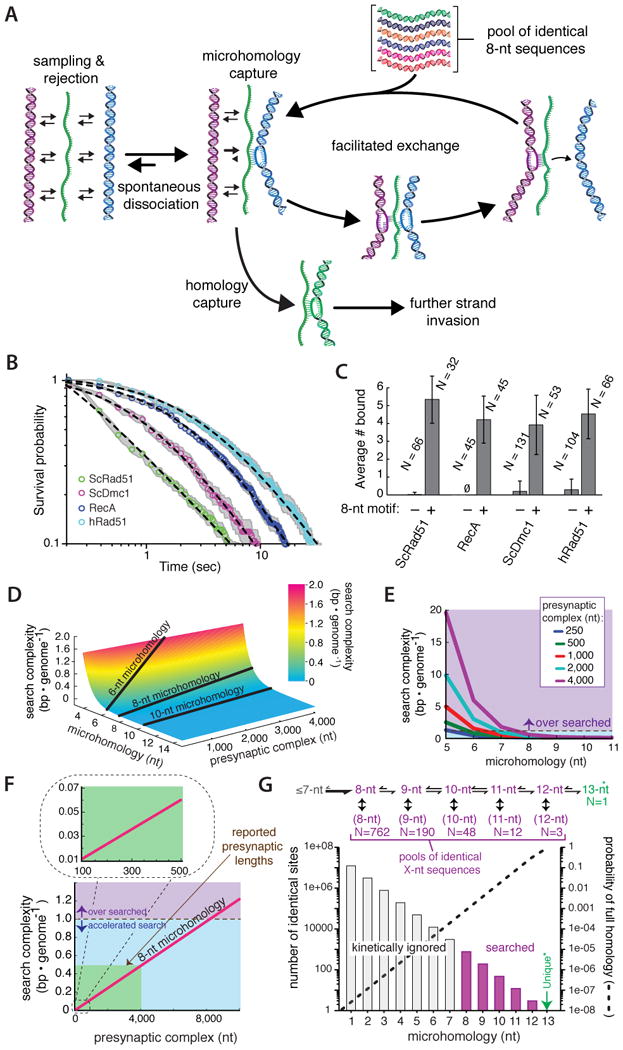
(A) Model depicting a homology search mechanism involving rapid sampling and rejection of DNA lacking microhomology, followed by eventual capture of an 8-nt tract of microhomology and facilitated exchange allowing for an iterative search through sequence space. Additional details are presented in the main text. Plots showing (B) power-law behavior during dsDNA sampling, and (C) microhomology-dependent binding for E. coli RecA, hRad51, and S. cerevisiae Dmc1; data presented for ScRad51 are reproduced from Figures 3B & 4E for comparison. The plus and minus 8-nt motif designations in (C) correspond to Atto565-DNA2.1 and Atto565-DNA2.0, respectively, and N corresponds to the number of PCs counted, and error bars represent S.D. (D) Surface plot showing how search complexity varies with PC length and the length of microhomology necessary for dsDNA interrogation. (E) Variation in search complexity for search models employing different lengths of microhomology, as indicated. (F) Relationship between search complexity and PC length for recognition involving 8-nt of microhomology. The green shaded region encompasses length estimates for S. cerevisiae PCs. (G) Fraction of the S. cerevisiae genome that can be kinetically ignored when employing a length-dependent search mechanism based on recognition of 8-nt motifs.
