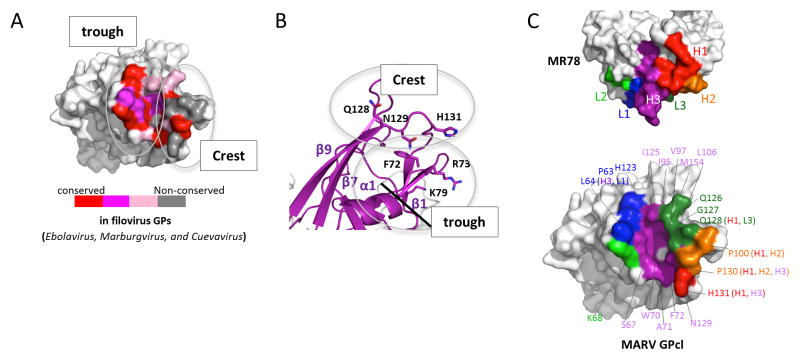
Figure 3. MR78 recognizes a conserved epitope at the apex of cleaved GP1.
(A) Conservation of the MR78 epitope among filovirus GPs, mapped onto one monomer of MARV GPcl. Sequence alignment was performed in ebolavirus (Ebola, Sudan, Reston, Taï Forest, Bundibugyo), marburgvirus (Musoke, Angola, Popp, Ci67, DRC1999, Ravn), and cuevavirus (Lloviu) genuses. Residues identical across the filoviruses are colored red; residues that possess strong similarity, magenta; weak similarity, pink; no similarity, gray. (B) The apex of cleaved MARV GP1, where Fab MR78 binds, forms a wave crest-and-trough morphology (magenta). The hydrophilic crest and the hydrophobic trough each contain residues previously shown to be critical for virus entry (Dube et al., 2009; Manicassamy et al., 2005; Manicassamy et al., 2007; Mpanju et al., 2006). The diagonal black line indicates the base of the trough. See also Figure S4. (C) Surface representation of the interface between one monomer of MARV GPcl (bottom) and Fab MR78 (top). CDR H1 is colored red; CDR H2, orange; CDR H3, purple; CDR L1, blue; CDR L2, green; CDR L3, forest green. The footprint on MARV GPcl is colored according to the CDR that mediates the contact. GP residues contacted by MR78 are indicated, and colored according to the CDR that mediates the contact (CDR names in parentheses).