Abstract
A combinatorial approach was used to present primary neurons with a large library of topographical features in the form of micropatterned substrate for high-throughput screening of physical neural-guidance cues that can effectively promote different aspects of neuronal development, including axon and dendritic outgrowth. Notably, the neuronal-guidance capability of specific features was automatically identified using a customized image processing software, thus significantly increasing the screening throughput with minimal subjective bias. Our results indicate that the anisotropic topographies promote axonal and in some cases dendritic extension relative to the isotropic topographies, while dendritic branching showed preference to plain substrates over the microscale features. The results from this work can be readily applied towards engineering novel biomaterials with precise surface topography that can serve as guidance conduits for neuro-regenerative applications. This novel topographical screening strategy combined with the automated processing capability can also be used for high-throughput screening of chemical or genetic regulatory factors in primary neurons.
The development and assembly of the nervous system not only contribute to topographically conserved areas with local specialization, but also globally integrated activity at many levels ranging from firing of a single neuron to overall behavioral output. In addition to various chemical signals that have been extensively reported1,2,3, the extracellular environment also presents to cells a variety of geometrically-defined, three dimensional physical cues or topographies, on the order of micro- and nano-scale, that significantly affect the development, function, and regeneration of nervous tissues4,5,6,7,8.
Specifically, neurons have been reported to be responsive to the local topography, with the earliest evidence presented by Paul Weiss in 19459. Neural tissue contains a complex three-dimensional structure with various topographical features spanning a large spectrum of morphologies and sizes10,11. In particular, neural cell bodies typically have diameters on the order of 10 μm (10–50 μm), whereas axons and dendrites have diameters in the micrometer range (typically, 0.2–3 μm)12; the filopodia of rat hippocampal neuron growth cones have been reported to have thicknesses in multiples of 60 nm13. Non-myelinated axons were qualitatively reported to have a smoother surface, while extracellular matrix (ECM) components like fibronectin, collagen, and laminin have sub 100 nm features14,15,16. There are a number of instances in the developing nervous system in which cells migrate along the tracts of other cells or oriented ECM fibers by contact guidance1,17. For example, migrating neuroblasts of the external granular layer of the cerebellum follow axon tracts that have already been laid18. However, compared to current understanding about the chemical neural guidance cues, the effects of topographical features on the development of nervous system, especially the distinct roles on the growth of axon or dendrites, are still remains elusive.
Recently, the application of micro- and nano-fabrication technologies in biomedical research has enabled creation of biomimetic structures that allow studying topographical regulation of cellular functions with unprecedented spatial and temporal resolution19,20,21. A significant milestone in this direction was the use of photolithography to pattern hydrophilic and hydrophobic compounds onto a silicon (Si) surface22, providing micrometer-level control over neuronal attachment and growth. Subsequently, a range of micropatterning techniques have evolved that enable defined placement of biomolecules on diverse substrates. Soft lithography, in particular, offers several advantages for the preparation of surfaces with cellular scale features. Recently, various patterned substrates, including micropatterns of guidance proteins23,24, electrospun fibers25,26, micro-/nano-structures27 and microbeads8, have been employed to study the neuronal response to microscale topography. Prominently, these topographical features have been utilized to inhibit the initiation of glial scarring, by altering the geometry, roughness, size, shape, and materials of neural implants and to study differentiation28,29. However, few systematic studies have been conducted to screen the precise topographical features that can promote or inhibit different aspects of neural functions30,31.
This study encompasses a combinatorial materiomics approach to identify physical neural-guidance cues; growing neuronal cells are subjected to a diverse set of topographical features in the form of micromolded substrates. In a high-throughput manner, we compare and analyze different aspects of neuronal development, including axon and dendritic outgrowth. Further, the neural-guidance effects derived from specific microscale topography patterns were automatically identified using in-house customized image processing algorithms, thus significantly increasing the analysis throughput with minimal subjective bias. Various patterns were designed for this study, including isotropic patterns (e.g. dots, grids, squares) that are symmetrical along both axes and anisotropic patterns (e.g. gratings) that are not symmetrical along both axes. The screening results indicate that the anisotropic topographies promote axonal extension, and some of the features also promote dendritic extension in comparison to isotropic topographies. Analysis of the neuronal dendritic branching indicates a preference for planar substrates rather than the microscale topographies. It is expected that the results from this work can be readily applied to engineer novel biomaterials with precise surface topography and creation of guidance conduits for neuro-regenerative applications. This novel screening strategy combined with automated software processing can also be used for in vitro high-throughput screening of chemical or genetic regulatory factors in cultured neurons.
Results
Effects of patterned topography on neuron growth
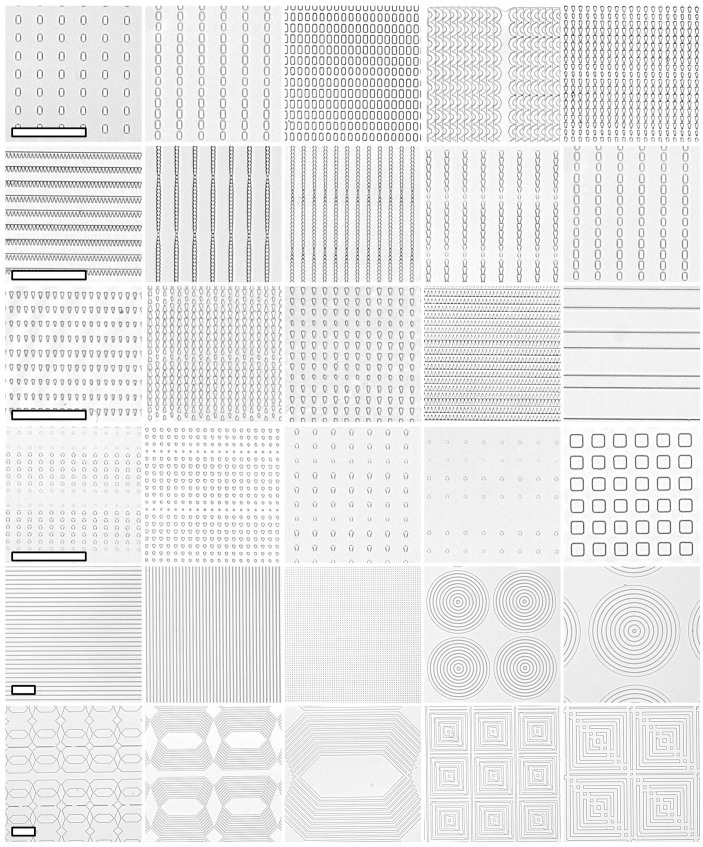
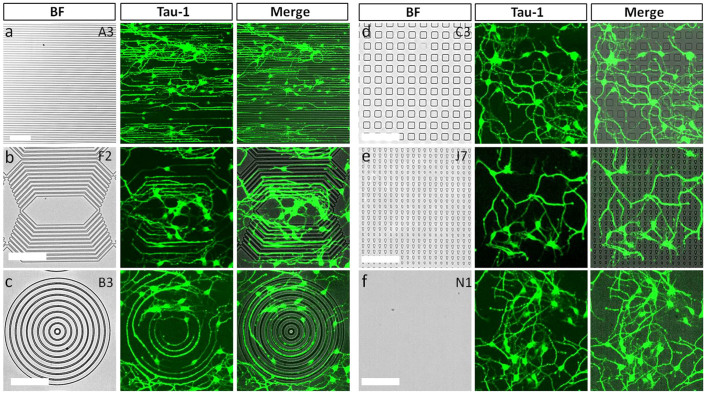
To systematically investigate the guidance effects by topography, we first designed and constructed a library of micrometer scale features ranging from 2 to 15 μm in size (Table 1). The feature geometries were either isotropic (dots, grids, and squares) or anisotropic (linear, circular, or angular gratings; triangles, and rectangles); planar substrate served as control, Figure 1 shows the brightfield images of some of the prominent features. These microscale features were 1 μm tall, and were replica molded with polydimethylsiloxane (PDMS) polymer. To evaluate the effects of patterned topography on axonal outgrowth, the primary hippocampal neurons were seeded and cultured for two days and then stained with axon specific markers (Tau1). At this stage, axons are already specified in a regular hippocampal culture32. As shown in Figure 2, axons from hippocampal neuron cells grew on different micropatterned topographical features, and were differently contact-guided. The guidance was most obvious on the continuous grating and circular patterns as shown in Figure 2a, b, and c, the guided axons were following the transition edge between ridges (features raised above substrate) and grooves (regions between ridges) in these patterns. Further, the axons were longer on the gratings with width of 5 and 15 μm as compared to the narrow width patterns, and irrespective of the constant interspacing that was 2 μm. However, only minor guidance effects were observed with sporadic or isolated features, such as the patterned dots and triangles shown in Figure 2 d and e. In this case the axons were only guided along a few dots or triangles before losing tracks after a short distance. Axons on a flat surface are also shown in Figure 2 f for comparison.
Table 1. List of microscale topography patterns used for high-throughput screening of neural guidance cues.
| Code | Patterns | Topography feature of patterns |
|---|---|---|
| A1-4 | Gratings | Straight grating with line width 2, 5, 10, 15 μm, space 2 μm |
| B1-4 | Circle | Circle with line width 2, 5, 10, 15 μm, space 5 μm |
| C1-4 | Dot | Dot with diameter 2, 5, 10, 15 μm |
| D1-3 | Grid | Grid with line width 2, 5, 10 μm, space 2, 5, 10 μm |
| E1-4 | Square | Square with line width 2, 5, 10, 15 μm, space 2, 5, 10, 15 μm |
| F1-3 | Angle | Angle with line width 2, 5, 10 μm, space 2, 5, 10 μm |
| H1-4 | Rectangle 1–4 | Rectangle with Y-space 5 μm, X-space 5, 10, 15, 25 μm |
| H5-8 | Rectangle 5–8 | Rectangle with Y-space 10 μm, X-space 5, 10, 15, 25 μm |
| H9-12 | Rectangle 9–12 | Rectangle with Y-space 15 μm, X-space 5, 10, 15, 25 μm |
| I1-3 | Triangle 1–3 | Triangle with Y-space 0 μm, X-space 5, 10, 20 μm |
| I4-7 | Triangle 4–7 | Triangle with Y-space 5 μm, X-space 0, 5, 10, 20 μm |
| I8-11 | Triangle 8–11 | Triangle with Y-space 10 μm, X-space 0, 5, 10, 20 μm |
| I12-15 | Triangle 12–15 | Triangle with Y-space 20 μm, X-space 0, 5, 10, 20 μm |
| J1-4 | Long triangle 1–4 | Long triangle with Y-space 0 μm, X-space 0, 5, 10, 20 μm |
| J5-8 | Long triangle 5–8 | Long triangle with Y-space 5 μm, X-space 0, 5, 10, 20 μm |
| J9-12 | Long triangle 9–12 | Long triangle with Y-space 10 μm, X-space 0, 5, 10, 20 μm |
| K1-2 | Directional triangle | Directional triangle with space 1, 5 μm |
| L1 | Semi-circle | Semi-circle with diameter 5 μm |
| M1 | Gradient line 1 | Gradient line with decreasing width |
| M2 | Gradient line 2 | Gradient line with decreasing width but increasing gap |
| N1 | Flat | Flat pattern |
Figure 1. Brightfield images of chosen microscale topography features.
Scale bar indicates 100 μm in each row.
Figure 2. Axonal outgrowth (2 DIV) on different microscale topographical patterns.
(a) Gratings with 10 μm ridge and 2 μm grooves, (b) angles with 5 μm ridge and 5 μm grooves, (c) circles with 10 μm ridge and 5 μm grooves, (d) 10 × 10 μm2 dots with 10 μm spacing, (e) triangles with 5 μm space in Y direction and 10 μm space in X direction, and (f) flat surface. Scale bar: 100 μm.
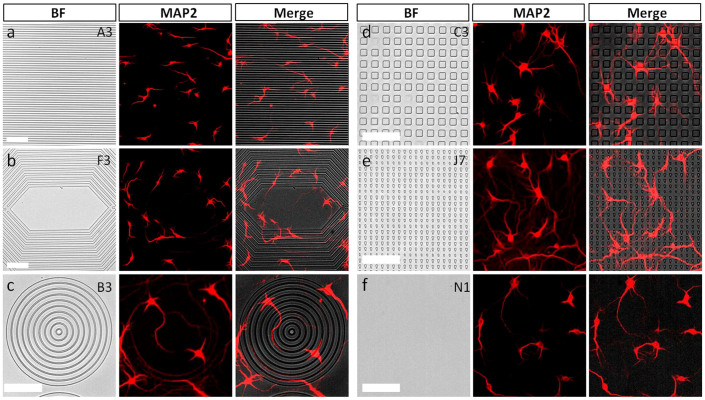
For further analysis, we maintained the neuronal cells in culture for 4 days, the time required for the cells to start developing dendrites. The results from the fluorescent staining of dendrites (MAP2) are shown in Figure 3. The imaging results showed that the topographical features were incapable of promoting the dendrite extension from the overlying neuronal cells, except for the directional triangular patterns that showed slightly better extension than other conditions. Image analysis results of the dendritic branching showed that there was no significant change in the number of branches on most of the microscale patterns, and the planar substrates showed slightly higher branching Figure 3f; indicating that the dendritic structures were unaffected by the topography.
Figure 3. Dendritic outgrowth (4 DIV) on microscale topographical patterns.
(a) Gratings with 10 μm ridge and 2 μm grooves, (b) angles with 10 μm ridge and 10 μm grooves, (c) circles with 10 μm ridge and 5 μm grooves, (d) 10 × 10 μm2 dots with 10 μm spacing, (e) triangles with 5 μm space in Y direction and 10 μm space in X direction, and (f) flat surface. Scale bar: 100 μm.
Automatic quantification of neuronal growth
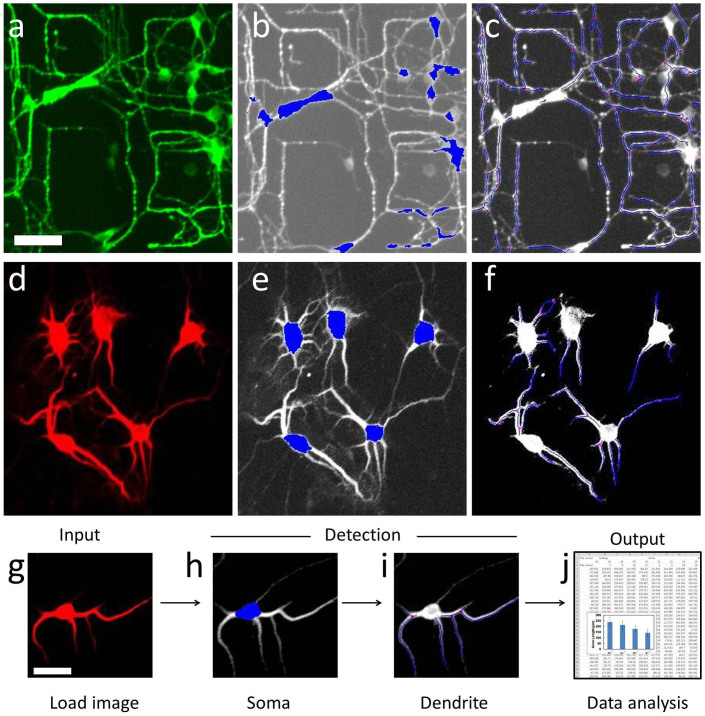
To quantitatively analyze the neuronal growth, we developed customized algorithms that could automatically quantify different aspects of neuronal growth. This Matlab-based program can be used to automatically capture delicate growth patterns, including axon length, dendrite length and arborization and neurite branching features, from large-amount image data. The automation significantly increased the throughput of our assays enabling us to accommodate more topographical guidance microstructures and test a large library in a high-throughput manner. Figure 4 shows an example of using the analysis scheme that was to analyze the axonal and dendritic outgrowth. The multiple steps employed during the analysis procedure are shown in Figure 4g–j, the detection algorithm also allowed identification of the cell bodies to calculate the neuronal cell density on each pattern. Figure 4a–c shows a representative result obtained during the analysis of axonal outgrowth. Figure 4d–f shows analysis of stained dendritic structures from neurons growing over a circular pattern, taken after the cells were cultured for 4 days. The blue lines indicate the length of the dendrite, whereas the red dots indicate the branching points.
Figure 4. Automated high-throughput analysis of neuronal guidance.
(a) Original axonal image of hippocampal neuron cells after culturing for 2 days on square pattern. (b) Cell body detection showing cell bodies in blue dots. (c) Axon length calculation showing axons as blue lines and branching points as red dots. (d) Original dendritic image of hippocampal neuron cells after culture for 4 days on gratings pattern. (e) Cell body detection showing cell bodies in blue dots. (f) Dendrite length and branches calculation showing dendrites as blue lines and branching points as red dots. (g) Original dendritic image of hippocampal neuron cell after culturing for 4 days. (h) Cell body detection showing cell body in blue dot. (i) Dendrite length and branches calculation showing dendrites as blue lines and branching points as red dots. (j) Output of data analysis. a–f scale bar: 50 μm; g–i scale bar: 30 μm.
High-throughput analysis of axonal and dendritic outgrowth
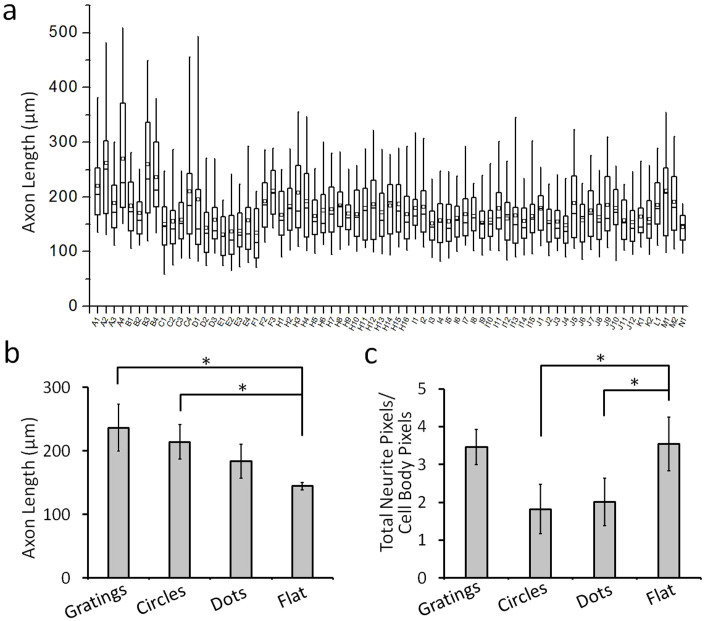
Proper axonal projection and dendritic arborization are critical for neural development as well as regeneration, it is thus helpful to guide and regulate the growth of axon. To implement our high-throughput analysis strategy using the customized program, we first analyzed the axonal length of neuronal cells (2 days in vitro (DIV)) growing on the patterned substrates with topographical features. As the analysis results indicate in Figure 5, the grating patterns were most effective at enhancing the axonal outgrowth. The average axon length of the neuronal cells on grating patterns was 237 ± 37 μm (mean ± s.e.m., same below), almost 60% longer than the neurons growing on flat plain substrates. For the circular patterns, the axon length was 214 ± 27 μm, 48% longer than the cells on the flat surface. On the breaking dot patterns, the axon length was much shorter (183 ± 27 μm), indicating that non-continuous dot patterns lead to shorter axon length.
Figure 5. Quantification and analysis of axonal outgrowth.
(a) Box-plot of the axonal length for cells growing on different patterns, n > 60 cells were quantified. The parts of the box plots indicate 25 and 75 percentiles, while the whiskers range is 5 and 95%. The square box indicates mean of the data, (n > 60 cells were quantified). (b) Average axonal length of neurons growing on gratings, circles, dots, and flat surface. (c) Value of total neurite pixel/cell body pixel of neurons growing on gratings, circles, dots, and flat surface. Error bars indicated s.e.m. from three independent experiments, *P < 0.05 by one way ANOVA test.
Our customized program enabled further comparison; the total pixels corresponding to an axon in a given image were normalized to the pixel number of cell bodies, to derive a total axonal outgrowth index. The results indicate that the axonal growth index is significantly larger on the grating pattern as compared to the circular and dot patterns (Figure 5c). Although the longest axon length for neurons on the flat surface was shorter than those growing on various patterns (isotropic as well as anisotropic), more branches of axon were observed in cells on the flat surface. Specifically for the grating pattern, the total axonal outgrowth index is similar to that on the flat surface, indicating that neurons grew longer but less branched axon on the grating pattern. In case of the circular and dot patterns, both the axon length and the branching were limited, which might be because of the space constraint, the small gap between the circles and similarly between the sporadic dots. These results support some of the observations from a previous study by Yang et al33, they have shown that the average total neurite length on a non-patterned substrate was longer than on substrates patterned with 10 μm wide features, and the patterns could restrict the cell nucleus and reduce neurite extension. Our results from the combinatorial topographical screening show that continuous anisotropic patterns such as gratings and circles provide strong guidance cue and promote directional axonal growth; but there is reduced axonal branching.
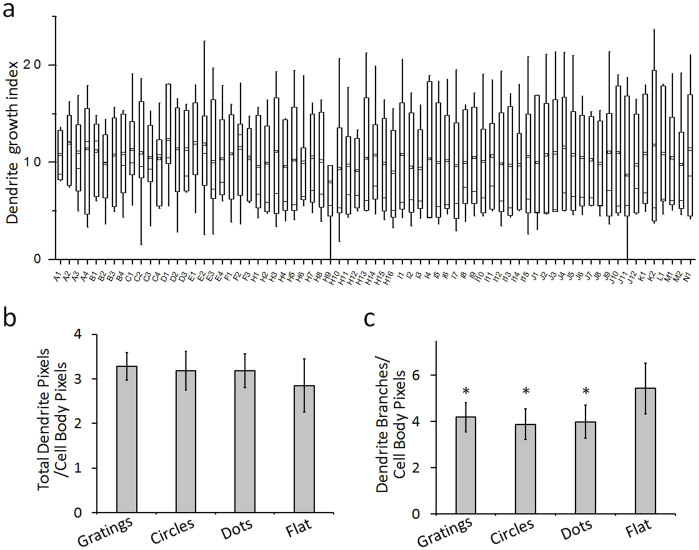
Correspondingly image processing and analysis were also performed on the dendrite stained neurons. The total dendritic pixels were normalized to cell body pixel to drive a dendritic growth index as shown in Figure 6. The larger the index number generally indicated a longer total dendrite length for cells on a specific pattern. The results showed that the dendritic growth was not sensitive to topographical features: the dendritic growth index on grating, circular, and dot patterns was 3.28 ± 0.31, 3.18 ± 0.43, and 3.19 ± 0.37, respectively, no significant difference was observed to that seen on flat surface (2.85 ± 0.59), as shown in Figure 6b and c. The branching index on the flat surface was higher compared to the patterned surface. Unlike axons, the dendritic growth was found to be insensitive on almost all topographical features tested in this study.
Figure 6. Quantification and analysis of dendritic outgrowth.
(a) Box-plot of dendrite growth index for cells growing on different patterns, n > 60 cells were quantified. The parts of the box plots indicate 25 and 75 percentiles, while the whiskers range is 5 and 95%. The square box indicates mean of the data, (n > 60 cells were quantified). (b) Dendritic growth index for neurons on patterns with gratings, circles, dots, and flat surface. The index was derived by taking ratio of total dendritic pixels to cell body pixels. (c) Dendrite branching index for neurons on patterns with gratings, circles, dots, and flat surface. The index was drived by taking the ratio of total branching points to cell body pixels. Error bars indicated s.e.m from three independent experiments, * P < 0.05 by one way ANOVA test.
Discussion
Novel features of the high-throughput neural-guidance topography chip
The topography chip for high-throughput screening of physical cues as implemented in this study demonstrates a novel concept that enables parallel investigation of multiple neuronal guidance aspects (e.g. length of neurites, branching). The conventional cell-topography studies are costly and labor-intensive given the limited number of topographies that can be tested in a study, and the cumbersome manual procedure for image analysis and data processing. We designed our platform to include 71 distinct geometric patterns onto two PDMS platforms, thus enabling high-throughput combinatorial topographical screening. To check the influence of regularly patterned sporadic structures and continuous guidance on neural guidance either isotropic (dots, grids, squares) or anisotropic (linear, circular, and angular gratings; triangles, semicircles) structures were designed. The procedure for making the substrates involves fabricating a pattern master and PDMS polymer molding, making it easy to implement and enabling fast test cycles.
The standard image processing software is not able to provide a comprehensive analysis of the different aspects of neuronal morphology, also performing the analysis manually is labor-intensive and prone to observer-bias. To address this issue, we developed algorithms that automated the analysis of multiple images with better precision. The program integrates two novel features that will aid in the analysis of neuronal guidance. First, it effectively identifies the neuronal bodies and can determine the cell density, as well as the axon and/or dendrite length. Second, the program can also measure the branching of the extended structure. This feature makes the image-processing step much faster and eliminates any user prone errors.
Topographical cues regulate neural guidance
Topographical regulation of neuronal cell morphology has been documented in literature5, Our results from combinatorial topographical screening show that continuous anisotropic patterns such as gratings and circles provide strong guidance cue and promote directional axonal growth; but there is reduced axonal branching. The anisotropic grating patterns were most effective at enhancing the axonal outgrowth. The average axon length of the neuronal cells growing on grating patterns was approximately 60% greater, and those on the circular patterns approximately 48% greater than the cells growing on flat planar substrates. Similarly, on the isolated sporadic dot patterns, the axon length was much shorter, indicating that isolated dot patterns lead to shorter axon length, compared to grating or circle patterns. The results from dendritic tracking indicate that the dendrites were insensitive to topographical features, and the branching index on all features was lower as compared to that on flat surface. Overall, the result showed that topographical patterns could be utilized not only to provide guidance cues but also to determine the differential sensitivity of the neurites.
This integrated platform with automated analysis can easily reduce the testing time for a small library of topographical features from 4–5 weeks as in case of a conventional platform, to less than a week, only limited by the time required for optimal neuronal outgrowth. This novel combinatorial screening strategy can also be employed for in vitro high-throughput screening applications for identifying chemical, genetic regulatory factors in neuronal cells and biophysical parameters driving differentiation.
Conclusion
This work introduces a novel, easy to implement platform that can be used for combinatorial screening of topographical features that act as guidance cues and promote neuronal-guidance. Performing topographical screening in traditional in vitro culture is challenging, the microscale topography chip was designed and developed to address this limitation. A microarray format was adopted, and different isotropic shapes as well as anisotropic patterns were chosen. A total of 71 features were designed to extensively cover the different combination of shapes, sizes, and spacing. The chip was fabricated using the traditional photolithography to fabricate the master mold in Si followed by micromolding of PDMS polymer substrate. In order to expedite the image analysis, with minimal bias, we developed customized software for measuring the cell density, neurite length, and branching. A systematic study was then conducted to discern the effect of the microstructured guidance cues on the growing neurons. The results indicate that topographical parameters play a crucial role in determining not only the length of the axons and dendrites, but also their branching. Specifically, we found that the anisotropic topographies promote both axonal extension and in some cases dendritic extension, relative to the isotropic topographies and planar substrate. Analysis of the branching showed that the dendritic branching index was higher on planar substrates. Overall, we have demonstrated that this novel topography chip enables identification of neuronal-guidance cues; these results will augment our understanding of design parameters that can be useful for developing better neural implants. Furthermore, this chip can easily be adapted for identifying chemical, genetic regulatory factors in neuronal cells, and aptly be used as a high-throughput platform for neuroscience research.
Methods
Fabrication of engineered cell culture platform
Engineered platforms for neuron growth were fabricated using Si molds and PDMS replicas. Patterns consisting of gratings, circles, and dots with 5–15 μm wide ridges and 1 μm deep grooves were fabricated in Si and transferred onto PDMS substrates. First, 1 μm photoresist (SPR6112B) was spin-coated onto a clean Si wafer, this was followed by standard contact-mode UV exposure for 10 s, a 60 nm thick chromium mask was used for the patterning. The unexposed photoresist was removed using a lift-off process in acetone. The patterned Si wafer was then etched to a depth of 1 μm using the reactive ion etching (RIE) process operated at 16/4 sccm CF4/O2, 20 mTorr chamber pressure, and 100 W rf power. In order to promote easy demoulding of the PDMS replica, the etched Si master mold was coated with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (FOTS) (Sigma-Aldrich, 97%), that renders the surface hydrophobic and facilitates demolding. A thin coating of FOTS was deposited onto the Si mold in a vacuum chamber for 5 h at 20°C. PDMS prepolymer (silicone elastomer base : curing agent w/w = 10:1, Dow Corning Sylgard 184) was poured onto the Si master mold in a culture dish and degassed for 15 min at 20°C, followed by annealing at 80°C for 5 h on a temperature controlled hot plate. Cured PDMS replica with 1–2 mm thickness was then gently peeled off from the Si mold, cleaned and stored until further use. The final dimension of the topography chip was 10 mm by 10 mm with 71 distinct patterns on two chips. The replication fidelity of the microstructures was determined using bright-field imaging on an optical microscope.
Cell culture
Hippocampal neurons were cultured on PDMS substrates with various microfabricated patterns. Prior to cell culture, the PDMS substrates were cleaned by ultrasonication in 70% ethanol for 5 min, and rinsed thoroughly with DI water. These PDMS substrates were coated with polylysine (Sigma-Aldrich) at 100 μg/ml following overnight incubation, and then with laminin at 10 μg/ml (Life Technologies) for 4 h, prior to seeding the neuronal cells.
As previously described34, hippocampi tissue were dissected from E18 Sprague Dawley rats, and treated with papain (Sigma) for 30 min at 37°C. Dissociated neurons were prepared by triturating enzymatic treated tissue with a 1 ml pipette tip in Dulbecco's modified eagle medium (DMEM, Life Technologies) solution contain 10% fetal bovine serum (FBS). Neurons were then seeded onto the coated PDMS substrates at a density of 2–3 × 104/cm2. After the initial adhesion of neuron cells (2 h after seeding), the medium was replaced by Neurobasal growth medium supplemented with B27 (Life Technologies), L-glutamine and penicillin/streptomycin. The non-adherent cells were gently washed off. The devices were maintained in a standard cell culture incubator at 37°C and 5% CO2 with half of the growth medium replaced with fresh medium every 3–4 days.
Immunocytochemistry
Neuronal cells cultured on PDMS substrates were washed with phosphate-buffered saline (PBS) and with fixed with 4% paraformaldehyde (w/v) for 30 min, permeablized by treatment with 0.25% Triton X-100 for 15 min, and then blocked with 4% bovine serum albumin (BSA) in PBS for 2 h at room temperature (or overnight at 4°C). Neuronal cells at 2 DIV were incubated for 2 h at room temperature with primary antibodies including mouse anti-Tau1 (Millipore) and chicken anti-Tubulin (Millipore) in 4% BSA that prevented nonspecific antibody binding. Neuronal cells at 4 DIV were incubated with primary antibodies including rabbit anti-MAP2 (Millipore) and chicken anti-Tubulin (Millipore) in 4% BSA for 2 h at room temperature. Following incubation with primary antibodies, the cultured neuronal cells were rinsed with PBS, and incubated with the secondary antibodies for 1 h, followed by rinsing with PBS.
Microscopy
Fluorescence images were acquired using an inverted microscope (IX81, Olympus) equipped with a CMOS camera (Neo, Andor), using a 10 × objective with a numerical aperture of 0.4. Each sample was scanned with an exposure time of 100 ms at a resolution 2560 × 2160 pixels. A montage of 142 images for each labeled antibody was acquired with the respective filter set. All of the scanned images were then stitched together into one large montage (17458 by 14658 pixels) and corrected for overlap, intensity.
Image analysis
To quantify the growth of axons and dendrites on patterned substrates, customized computer algorithm were developed that enabled image analysis and automated measurement of the neurite length and branching numbers. A cell body detection algorithm was also developed to obtain the cell density on each patterned substrate and enabled calculating the variation. More details about the algorithms and the processing procedures explained in the Supplementary Note S1. The source code of the program is included in Supplementary Note S2.
For the cell body detection algorithm, we first set a threshold to segment the original image to a binary image. We then used morphological filter to remove small area pixels, classify the connected components, and differentiate the cell bodies. Finally, to perform classification we used equivalent circle to remove the unrelated pixels and obtain the cell body image with the cell bodies identified.
For the length calculation algorithm, we first set a threshold to segment the original image to a binary image. We then performed morphological skeletonization to reduce the width of the dendrite to a single-pixel width. The small single-pixel lines were removed from the cell body, and the length of axon or dendrite was obtained by the total pixel accumulation over the single-pixel lines. For the branch calculation algorithm, we detected the corners of the dendrite paths used in the image from the length calculation. The number of dendrite branches was obtained by the accumulation of the total number of corners.
Author Contributions
P.S. and S.W.P. conceived the project and wrote the manuscript. W.L., Q.T., A.N. and W.Q. conducted the experiments and analyzed the data. A.D.J. and W.Q. also contributed to the writing of the manuscript.
Supplementary Material
Supplementary Information
SI - Source code
Acknowledgments
The authors would like to acknowledge the financial support from the Center for Biosystems, Neuroscience, and Nanotechnology of City University of Hong Kong under project number 9360148. This work was also supported by the University Grants Council of Hong Kong (ECS125012, GRF11211314 and GRF 9041907), National Science Foundation of China (81201164), and grants from the City University of Hong Kong (7200269, 9610215, 9667077, 7003022, 9360148, and 9380062).
References
- Guan K. L. & Rao Y. Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci 4, 941–956 (2003). [DOI] [PubMed] [Google Scholar]
- Shi P. et al. Synapse microarray identification of small molecules that enhance synaptogenesis. Nature Communications 2, 510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Nedelec S., Wichterle H. & Kam L. C. Combined microfluidics/protein patterning platform for pharmacological interrogation of axon pathfinding. Lab Chip 10, 1005–1010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P., Shen K., Ghassemi S., Hone J. & Kam L. C. Dynamic Force Generation by Neural Stem Cells. Cell Mol Bioeng 2, 464–474 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Kim D., Mitchel J. A. & Bellamkonda R. V. Topography, cell response, and nerve regeneration. Annual review of biomedical engineering 12, 203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranes K., Chejanovsky N., Alon N., Sharoni A. & Shefi O. Topographic cues of nano-scale height direct neuronal growth pattern. Biotechnol Bioeng 109, 1791–1797 (2012). [DOI] [PubMed] [Google Scholar]
- Turunen S., Haaparanta A. M., Aanismaa R. & Kellomaki M. Chemical and topographical patterning of hydrogels for neural cell guidance in vitro. J Tissue Eng Regen Med 7, 253–270 (2013). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Fabrication of density gradients of biodegradable polymer microparticles and their use in guiding neurite outgrowth. Advanced functional materials 20, 1632–1637 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P. The problem of specificity in growth and development. Yale J Biol Med 19, 235–278 (1947). [PMC free article] [PubMed] [Google Scholar]
- Moore S. W. & Sheetz M. P. Biophysics of substrate interaction: influence on neural motility, differentiation, and repair. Dev Neurobiol 71, 1090–1101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A., Moriguchi T. & Salem N. Decrease in neuron size in docosahexaenoic acid-deficient brain. Pediatr Neurol 26, 210–218 (2002). [DOI] [PubMed] [Google Scholar]
- Hollenbeck P. J. & Bamburg J. R. Comparing the properties of neuronal culture systems: a shopping guide for the cell biologist. Methods Cell Biol 71, 1–16 (2003). [DOI] [PubMed] [Google Scholar]
- Parpura V., Haydon P. G. & Henderson E. Three-dimensional imaging of living neurons and glia with the atomic force microscope. J Cell Sci 104 (Pt 2), 427–432 (1993). [DOI] [PubMed] [Google Scholar]
- Wess T. J. Collagen fibril form and function. Adv Protein Chem 70, 341–374 (2005). [DOI] [PubMed] [Google Scholar]
- Pompe T., Renner L. & Werner C. Nanoscale features of fibronectin fibrillogenesis depend on protein-substrate interaction and cytoskeleton structure. Biophys J 88, 527–534 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K., Hunter I. & Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J 4, 148–160 (1990). [DOI] [PubMed] [Google Scholar]
- Gil-Sanz C. et al. Cajal-Retzius Cells Instruct Neuronal Migration by Coincidence Signaling between Secreted and Contact-Dependent Guidance Cues. Neuron 79, 461–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O., Patel R. & Miller R. H. Migration of neuroblasts along preexisting axonal tracts during prenatal cerebellar development. J Neurosci 6, 867–876 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Tsai J., Shi P. & Kam L. C. Self-aligned supported lipid bilayers for patterning the cell-substrate interface. J Am Chem Soc 131, 13204–13205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q. et al. Influence of engineered surface on cell directionality and motility. Biofabrication 6, 015011 (2014). [DOI] [PubMed] [Google Scholar]
- Li W. et al. NeuroArray: a universal interface for patterning and interrogating neural circuitry with single cell resolution. Sci Rep 4, 4784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D., Kahler K. H. & Hockberger P. E. Controlled outgrowth of dissociated neurons on patterned substrates. J Neurosci 8, 4098–4120 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez N., Lu Y., Chen S. & Schmidt C. E. Immobilized nerve growth factor and microtopography have distinct effects on polarization versus axon elongation in hippocampal cells in culture. Biomaterials 28, 271–284 (2007). [DOI] [PubMed] [Google Scholar]
- Shi P., Shen K. & Kam L. C. Local presentation of L1 and N-cadherin in multicomponent, microscale patterns differentially direct neuron function in vitro. Dev Neurobiol 67, 1765–1776 (2007). [DOI] [PubMed] [Google Scholar]
- Christopherson G. T., Song H. & Mao H.-Q. The influence of fiber diameter of electrospun substrates on neural stem cell differentiation and proliferation. Biomaterials 30, 556–564 (2009). [DOI] [PubMed] [Google Scholar]
- Yang F., Murugan R., Wang S. & Ramakrishna S. Electrospinning of nano/micro scale poly (L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials 26, 2603–2610 (2005). [DOI] [PubMed] [Google Scholar]
- Migliorini E. et al. Acceleration of neuronal precursors differentiation induced by substrate nanotopography. Biotechnology and bioengineering 108, 2736–2746 (2011). [DOI] [PubMed] [Google Scholar]
- Dowell-Mesfin N. et al. Topographically modified surfaces affect orientation and growth of hippocampal neurons. Journal of neural engineering 1, 78 (2004). [DOI] [PubMed] [Google Scholar]
- Grill W. M., Norman S. E. & Bellamkonda R. V. Implanted neural interfaces: biochallenges and engineered solutions. Annu Rev Biomed Eng 11, 1–24 (2009). [DOI] [PubMed] [Google Scholar]
- Johansson F., Carlberg P., Danielsen N., Montelius L. & Kanje M. Axonal outgrowth on nano-imprinted patterns. Biomaterials 27, 1251–1258 (2006). [DOI] [PubMed] [Google Scholar]
- Hanson J. N. et al. Textural guidance cues for controlling process outgrowth of mammalian neurons. Lab on a Chip 9, 122–131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G. & Goslin K. Culturing Nerve. Cells MIT Press, 1998. [Google Scholar]
- Cao H. et al. The effects of nanofiber topography on astrocyte behavior and gene silencing efficiency. Macromol Biosci 12, 666–674 (2012). [DOI] [PubMed] [Google Scholar]
- Kaech S. & Banker G. Culturing hippocampal neurons. Nat Protoc 1, 2406–2415 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
SI - Source code