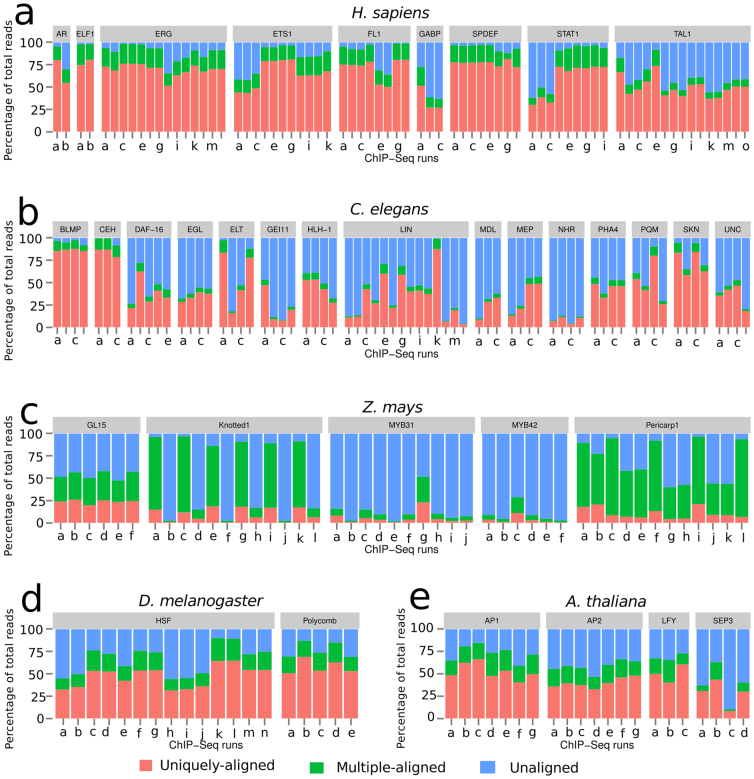
Figure 2. Proportions of uniquely-aligned, multiple-aligned, and unaligned reads in ChIP-Seq datasets of five organisms.
Each bar represents a ChIP-Seq run; several runs constitute an experiment for determining binding patterns of one transcription factor. Runs have been grouped into their respective TFs indicated in the gray boxes; each run has been coded by an identifier representing NCBI Short Read Archive accession number for a ChIP-Seq run (See Supplementary File 1 for the accession numbers corresponding to each identifier in the figure).