Abstract
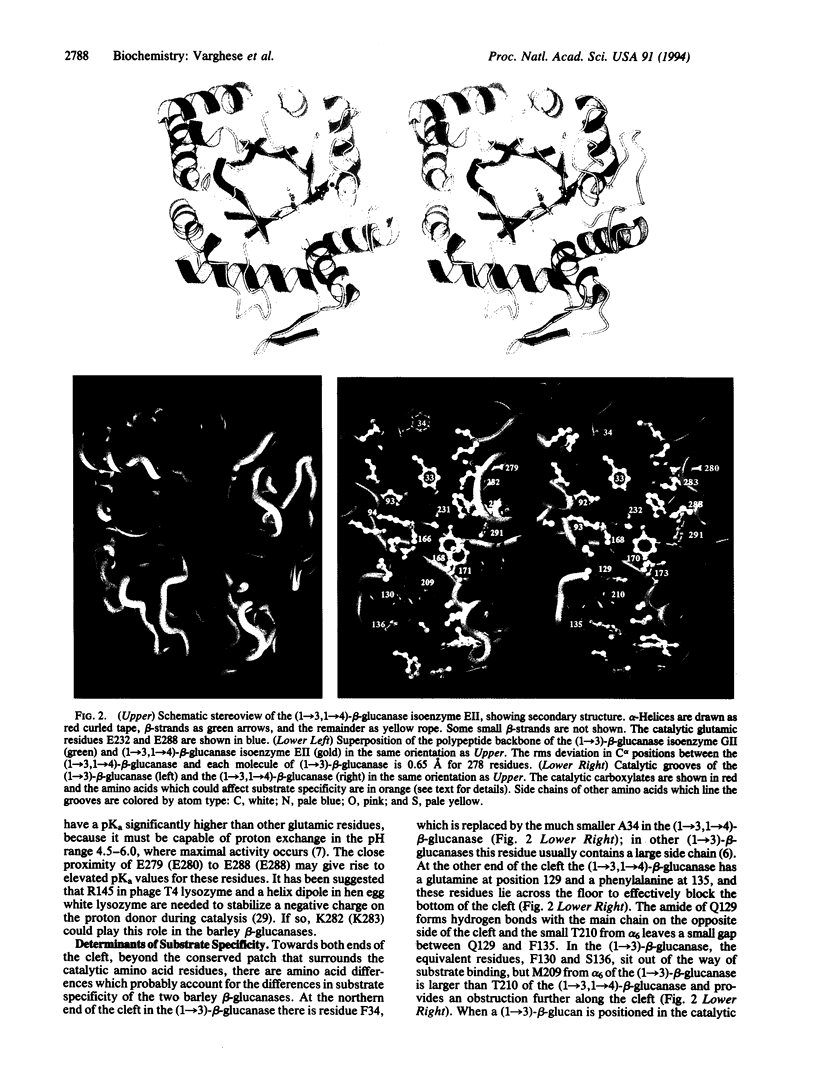
The three-dimensional structures of (1-->3)-beta-glucanase (EC 3.2.1.39) isoenzyme GII and (1-->3,1-->4)-beta-glucanase (EC 3.2.1.73) isoenzyme EII from barley have been determined by x-ray crystallography at 2.2- to 2.3-A resolution. The two classes of polysaccharide endohydrolase differ in their substrate specificity and function. Thus, the (1-->3)-beta-glucanases, which are classified amongst the plant "pathogenesis-related proteins," can hydrolyze (1-->3)- and (1-->3,1-->6)-beta-glucans of fungal cell walls and may therefore contribute to plant defense strategies, while the (1-->3,1-->4)-beta-glucanases function in plant cell wall hydrolysis during mobilization of the endosperm in germinating grain or during the growth of vegetative tissues. Both enzymes are alpha/beta-barrel structures. The catalytic amino acid residues are located within deep grooves which extend across the enzymes and which probably bind the substrates. Because the polypeptide backbones of the two enzymes are structurally very similar, the differences in their substrate specificities, and hence their widely divergent functions, have been acquired primarily by amino acid substitutions within the groove.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., Corran P. H., Furth A. J., Milman J. D., Offord R. E. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975 Jun 19;255(5510):609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- Brünger A. T. Crystallographic refinement by simulated annealing. Application to a 2.8 A resolution structure of aspartate aminotransferase. J Mol Biol. 1988 Oct 5;203(3):803–816. doi: 10.1016/0022-2836(88)90211-2. [DOI] [PubMed] [Google Scholar]
- Chen L., Fincher G. B., Høj P. B. Evolution of polysaccharide hydrolase substrate specificity. Catalytic amino acids are conserved in barley 1,3-1,4- and 1,3-beta-glucanases. J Biol Chem. 1993 Jun 25;268(18):13318–13326. [PubMed] [Google Scholar]
- Chen L., Garrett T. J., Varghese J. N., Fincher G. B., Høj P. B. Crystallization and preliminary X-ray analysis of (1,3)- and (1,3;1,4)-beta-D-glucanases from germinating barley. J Mol Biol. 1993 Dec 5;234(3):888–889. doi: 10.1006/jmbi.1993.1635. [DOI] [PubMed] [Google Scholar]
- Davies G. J., Dodson G. G., Hubbard R. E., Tolley S. P., Dauter Z., Wilson K. S., Hjort C., Mikkelsen J. M., Rasmussen G., Schülein M. Structure and function of endoglucanase V. Nature. 1993 Sep 23;365(6444):362–364. doi: 10.1038/365362a0. [DOI] [PubMed] [Google Scholar]
- Doan D. N., Fincher G. B. Differences in the thermostabilities of barley (1----3,1----4)-beta-glucanases are only partly determined by N-glycosylation. FEBS Lett. 1992 Sep 14;309(3):265–271. doi: 10.1016/0014-5793(92)80786-g. [DOI] [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Howard A. J., Nielsen C., Xuong N. H. Software for a diffractometer with multiwire area detector. Methods Enzymol. 1985;114:452–472. doi: 10.1016/0076-6879(85)14030-9. [DOI] [PubMed] [Google Scholar]
- Hrmova M., Fincher G. B. Purification and properties of three (1-->3)-beta-D-glucanase isoenzymes from young leaves of barley (Hordeum vulgare). Biochem J. 1993 Jan 15;289(Pt 2):453–461. doi: 10.1042/bj2890453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høj P. B., Hartman D. J., Morrice N. A., Doan D. N., Fincher G. B. Purification of (1-->3)-beta-glucan endohydrolase isoenzyme II from germinated barley and determination of its primary structure from a cDNA clone. Plant Mol Biol. 1989 Jul;13(1):31–42. doi: 10.1007/BF00027333. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Keitel T., Simon O., Borriss R., Heinemann U. Molecular and active-site structure of a Bacillus 1,3-1,4-beta-glucanase. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5287–5291. doi: 10.1073/pnas.90.11.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990 Jul 27;249(4967):380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- Slakeski N., Baulcombe D. C., Devos K. M., Ahluwalia B., Doan D. N., Fincher G. B. Structure and tissue-specific regulation of genes encoding barley (1----3, 1----4)-beta-glucan endohydrolases. Mol Gen Genet. 1990 Dec;224(3):437–449. doi: 10.1007/BF00262439. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., James M. N. Lysozyme revisited: crystallographic evidence for distortion of an N-acetylmuramic acid residue bound in site D. J Mol Biol. 1991 Jul 20;220(2):401–424. doi: 10.1016/0022-2836(91)90021-w. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Woodward J. R., Fincher G. B. Purification and chemical properties of two 1,3;1,4-beta-glucan endohydrolases from germinating barley. Eur J Biochem. 1982 Jan;121(3):663–669. doi: 10.1111/j.1432-1033.1982.tb05837.x. [DOI] [PubMed] [Google Scholar]
- Xu P., Wang J., Fincher G. B. Evolution and differential expression of the (1-->3)-beta-glucan endohydrolase-encoding gene family in barley, Hordeum vulgare. Gene. 1992 Oct 21;120(2):157–165. doi: 10.1016/0378-1119(92)90089-8. [DOI] [PubMed] [Google Scholar]