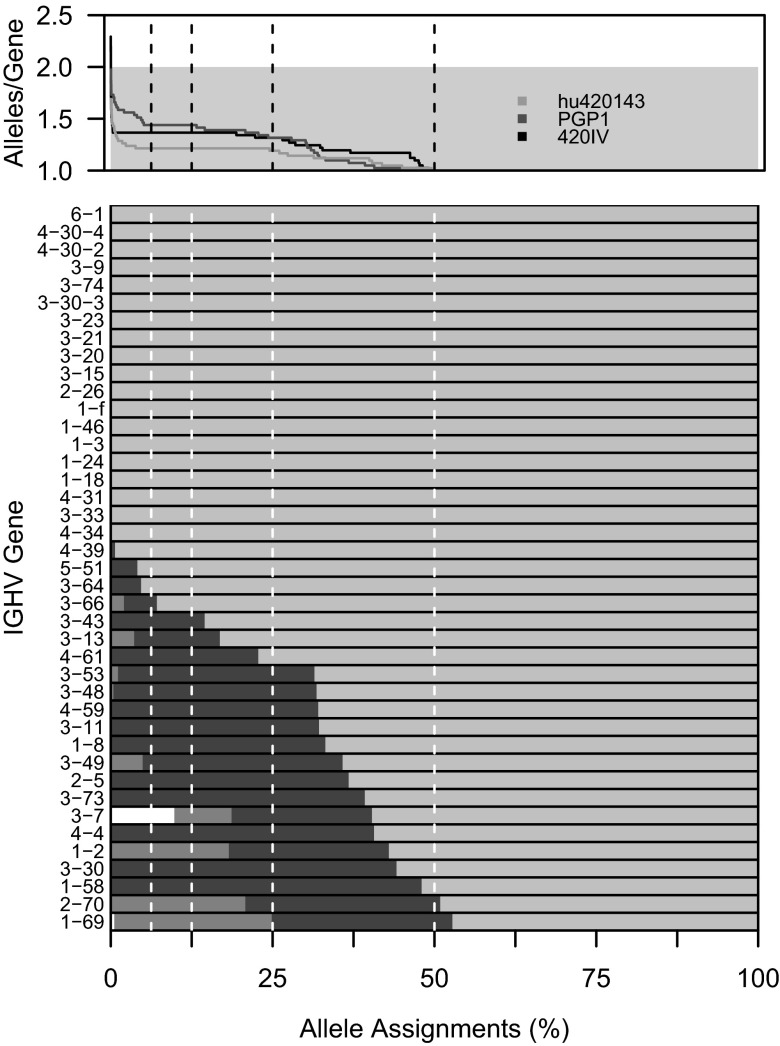
Fig. 6.
The influence of allele assignment frequency cut-offs on IGHV genotype zygocity. TIgGER was used to determine subject-specific IGHV genotypes using different values for the allele assignment frequency cut-off (i.e., fraction of assignments to a gene segment that are required to be composed of a single allele to be included in the genotype). For each of the three 454 datasets, the number of alleles included in the inferred IGHV genotype is shown as a function of the allele assignment frequency cut-off (Upper). For PGP1, the distribution of allele assignments is shown for each gene included in the inferred genotype (Lower). For each bar, the lightest gray represents the most common allele, darkest gray the second most common, medium gray the third, and white for all others.