Abstract
Objectives:
ALK-EML4 translocation is an established driver aberration in non-small cell lung cancer (NSCLC), with reported predilection for cases with signet ring histology. We assessed the presence of anaplastic lymphoma kinase (ALK) gene rearrangements in signet ring cancers arising in the stomach and colon.
Methods:
Histologically confirmed cases of signet ring adenocarcinoma of the stomach or the colon were identified. The presence of the classic ALK and EML4 fusion gene was initially determined by fluorescence in-situ hybridization (FISH) technique. Immunohistochemistry (IHC) was performed using two previously validated antibodies, ALK1 clone (1:100; DAKO) and 5A4 (Novocastra, Leica Biosystems) along with positive controls of ALK-translocated lung cancer.
Results:
We employed 42 cases of signet ring carcinoma diagnosed between 2001 and 2011; 25 gastric and 17 colon cancer. Median age 63.3 years; male/female 17/25; race, black 47.5%, white 47.5%, others, 5%; stage I, 21.4%; stage II, 31%; stage III, 26.2%; stage IV, 21.4%. One of 42 cases (2.3%) was positive for ALK translocation by FISH using the standard criteria of at least 15% positive cells for the break-apart signal (50–70 cells enumerated per case). Using a less restrictive cut-off of 10% positive cells, 7 cases (16%) were considered possibly positive. None of the ‘possibly positive’ cases was found to harbor ALK translocation by another molecular testing approach (IHC). IHC with two previously validated monoclonal antibodies showed 0 of 42 (0%) cases positive.
Conclusions:
ALK gene rearrangement is very rare in gastrointestinal cancers and enrichment strategy focusing on signet ring cell histology did not significantly improve the detection rate.
Keywords: ALK rearrangement, ALK-EML4 fusion, colon, signet ring cell cancer, stomach
Introduction
Colorectal cancer (CRC) and gastric cancers are leading causes of cancer-related mortality in the United States and worldwide [Ferlay et al. 2010]. The mortality arising from CRC or gastric cancer is due mainly to metastatic disease, which is not effectively controlled with currently available systemic therapies [Tsujii et al. 1997]. The traditional approach to drug development where agents are developed for clinically selected patient group based on tissue or site of origin [Park et al. 2004] is beginning to give way to targeted drug development for molecularly defined patient groups. This approach has been made possible by the improved understanding of the principal molecular mechanisms driving cancer development. As such, agents are now being developed to target patient subgroups with well-validated driver molecular aberration. The development of crizotinib for non-small cell lung cancer (NSCLC) harboring a translocation between the anaplastic lymphoma kinase (ALK) and echinoderm microtubule associated protein like 4 (EML4) genes is a clear example of this evolving paradigm in cancer therapeutics [La Thangue and Kerr, 2011; Roskoski, 2013]. The initial recognition of EML4-ALK rearrangement as a driver aberration in lung cancer was made in cell lines [Soda et al. 2007]. The subsequent confirmation of this finding in patient samples from various centers in Asia and the United States established the clinical relevance, leading to clinical development of specific agents against this target. The identification of a subset of patients with NSCLC harboring ALK gene translocation facilitated the successful development of crizotinib as a new treatment option in lung cancer [Ou et al. 2011].
Moreover, the unique clinical and pathologic features of ALK-rearranged lung cancer enabled case identification. For instance, ALK positive lung cancers are more likely to be diagnosed in younger age group (<50 years) and patients with no significant history of prior tobacco exposure [Galetta et al. 2012]. ALK rearranged lung cancer were also observed to be of mucinous histology and to have the signet ring morphology in some of the reported studies [Rodig et al. 2009; Jokoji et al. 2010; Popat et al. 2012]. There is no known anatomic or biologic mechanism to suggest that ALK gene rearrangement is selective or restricted to lung cancers. Therefore, attempts have been made to characterize other tumor types for the presence of ALK rearrangement as a prelude to testing ALK inhibitors in these tumor types [Grob et al. 2012; Krishnamurthy et al. 2013; Niu et al. 2013].
The incidence of ALK gene alterations in other malignancies such as colorectal or gastric cancer has not been well studied. Two prior studies reported a very low prevalence of EML4-ALK rearrangement in colorectal cancer of 0% (0 of 770) [Bavi et al. 2013] and 2.4% (2 of 83 patients) [Lin et al. 2009]. These studies, however, did not employ any enrichment strategy to improve the detection rate. We therefore hypothesized that the use of tumor histology as an enrichment strategy will lead to a higher rate of detection and facilitate the development of therapeutic agents for this subset of patients. We therefore decided to evaluate for EML4-ALK translocation in the subset of colon and gastric cancer with signet ring histology. In addition, we employed two different testing platforms to detect dysregulated ALK signaling, which in addition to the well-characterized fusion with EML4, can also result from specific activating mutations, gene amplification or fusion with other partner genes [Mosse et al. 2009].
Materials and methods
Patient samples
We reviewed cases of gastric and colorectal cancers treated at Winship Cancer Institute of Emory University between 2001 and 2011 in order to identify eligible cases for the study. The study was approved by the Emory Institution Review Board. A convenient sampling method was used whereby all cases of signet ring adenocarcinoma meeting the stated eligibility criteria during the period under review were included. The salient criteria for inclusion were: a pathologically confirmed diagnosis, availability of adequate tissue samples and reliable records for clinical care and survival. Electronic records of the selected cases of gastric and colorectal cancers treated were reviewed in order to obtain relevant clinical data including: demographics (age, gender, and ethnicity), date of diagnosis, type of treatment and course of disease, family or genetic predisposition, and other potential risk factors. The corresponding paraffin-embedded tissue specimens (FFPE) obtained at the time of original diagnosis were retrieved from the archives of the department of pathology. A total of 90% of the archival samples were suitable for inclusion.
Fluorescence in-situ hybridization Assay
The presence of the classic ALK-EML4 fusion gene was initially determined using the current gold standard break-apart fluorescence in-situ hybridization (FISH) test as described previously [Ou et al. 2011]. Interphase molecular cytogenetic studies using a commercially available ALK probe (Vysis LSI ALK Dual Color, Break Apart Rearrangement Probe; Abbott Molecular, Abbott Park, IL) were performed on paraffin-embedded sections that were deparaffinized three times in CitriSolv for 10 minutes each, dehydrated twice in 100% ethylic alcohol for 5 minutes. Tissue sections were then transferred to 85°C standard saline citrate (2xSSC) for 15 minutes, and protein was digested with 0.6 mg/ml Proteinase K (Sigma) at 45ºC for 16 minutes. After brief washing in 1x phosphate-buffered saline, the slides were sequentially dehydrated in alcohol (70%, 85%, and 100%) and air dried at room temperature. ALK probe was applied to the selected tumor area, which was covered and sealed with rubber cement. Slides were denatured at 85°C for 15 minutes, and probe hybridization was carried out overnight in a humidified chamber at 37°C. Tissue sections were washed in 0.3% NP40/1x SSC at 74°C for 2 minutes and then washed in 2xSSC at room temperature for 2 minutes. Slides were mounted in VECTASHIELD Mounting Medium (Vector Laboratories, Burlingame, CA) with 1.5 g/ml of 4, 6-diamidino-2-phenylindole. In this test, 5’ (red) and 3’ (green) fluorescent probes bind to areas upstream and downstream of the rearrangement breakpoint in exon 20 of the ALK gene(19).
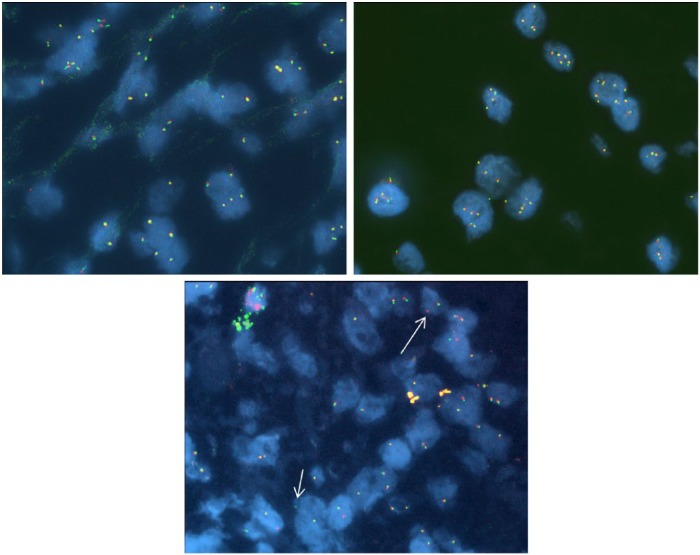
When the ALK gene is intact, the red and green signals are closely apposed together and appear fused. They are referred to as a fused signal, sometimes registering as a composite yellow signal (Figure 1). With ALK gene rearrangement, the signals move apart from each other to give the split signal, which is considered positive. We scored a total of 50 to 80 tumor cells per specimens and presence of at least 15% of cells showing separated 3’ALK and 5’ALK signals or single 3’ALK signals classified the specimen as positive for ALK rearrangement [Kwak et al. 2010].
Figure 1.
Break-apart fluorescence in-situ hybridization (FISH) probe for anaplastic lymphoma kinase (ALK): (1) ALK FISH break-apart probe set showing normal fused red/green signals; (2) ALK FISH break-apart probe set showing high number of copies of fused red/green signals per nucleus; (3) ALK FISH break-apart probe set showing isolated 3’ALK (single red signal - long arrow) and 5’ ALK (single green signal - short arrow).
Immunohistochemistry
Expression of ALK protein was determined using standard IHC techniques along with positive and negative controls. The commercially available antibodies employed for IHC were previously employed by other groups to determine ALK protein expression in pathologic specimens [Mino-Kenudson et al. 2010]. Briefly, following deparaffinization, antigen retrieval was performed using Target Retrieval Solution diluted 1:10 (pH 6). The slides were placed in this solution and heated to 120°C for 4 minutes using Biocare Decloaking Chamber. Afterward, the slides were allowed to cool in hot solution for 20 minutes, and were gently washed in running tap water. The slides were placed in a wash buffer bath (Tris Buffered Saline [TBS] and Tween 20), and endogenous peroxidase blocking was established with 3% H2O2 for 5 minutes. The slides were subsequently rinsed in TBS before incubation with primary antibodies ALK1 clone (1:100; DAKO) and ALK Clone 5A4 (Novocastra, Leica Biosystems). Protein expression was assessed jointly by two of the authors (GS; TKO) using previously published standard algorithm: negative staining (0), weak intensity (1+), moderate intensity (2+) and strong intensity (3+). The proportion of the staining cells were also determined and semi-quantitatively expressed as 0 (0%), 1+ (1–25%), 2+ (26–50%), 3+ (51–75%), 4+ (76–100%). Any sample with staining intensity ⩾2+ was considered positive for dysregulated ALK signaling.
Statistical analysis
The frequency of ALK dysregulation in gastrointestinal cancer is currently unknown. However, based on the NSCLC data, a 10% prevalence rate was considered interesting enough to pursue further development of a diagnostic or therapeutic intervention. Descriptive statistics such as mean, median, range and standard deviation of the mean score were generated for the histologic results. Inferential statistics were performed as applicable using Student’s t-test or Mann–Whitney test depending on the distribution of the data. The significance levels were set at 0.05 for all tests. The SAS statistical package V9.3 (SAS Institute, Inc., Cary, NC) was used for data managements and analyses.
Results
Clinical characteristics
We identified 42 cases of signet ring carcinoma from the cancer registry database; 25 gastric and 17 colon cancers. Table 1 details the baseline demographics and disease characteristics of the identified cases. The median age was 63.3 years with a female-to-male ratio of 1.5. There were equal numbers of African Americans and non-Hispanic white (47.5% each).
Table 1.
Patient and tumor characteristics.
| N = 42 | % | ||
|---|---|---|---|
| Gender | Female | 25 | 59.5 |
| Male | 17 | 40.5 | |
| Race | African American | 19 | 47.5 |
| Other | 2 | 5.0 | |
| White | 19 | 47.5 | |
| Missing | 2 | – | |
| Histology | Colon | 17 | 40.5 |
| Gastric | 25 | 59.5 | |
| Stage | I | 9 | 21.4 |
| II | 13 | 31.0 | |
| III | 11 | 26.2 | |
| IV | 9 | 21.4 | |
| Age | Overall | ||
| Mean (SD) | 63.2 (12.8) | ||
| Median (IQR) | 63.3 (5) |
SD, standard deviation; IQR, interquartile range.
ALK rearrangement detection by FISH
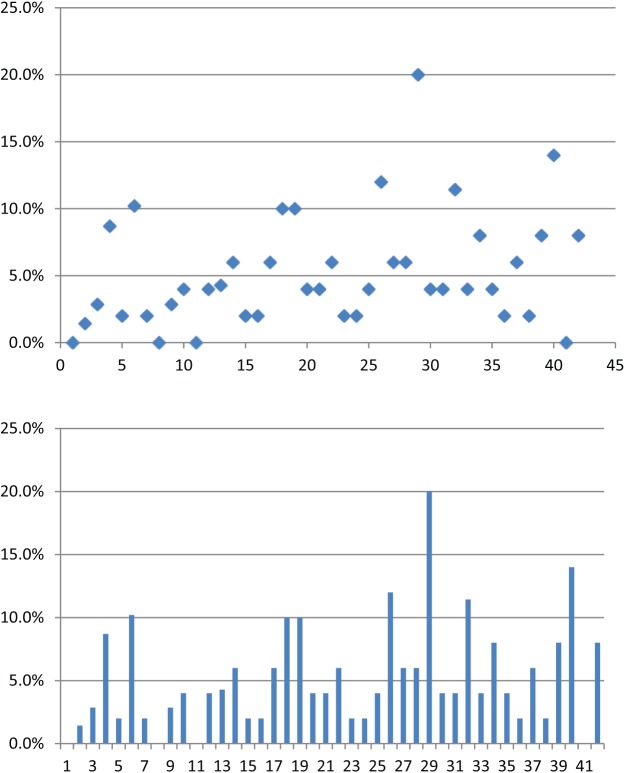
There was a single case (gastric adenocarcinoma with signet ring features) out of the 42 cases (2.3%) that was positive for ALK rearrangement by FISH, using the standard criteria of ⩾15% of cells being positive for the break-apart signal (Figure 2). This was a total gastrectomy specimen and evaluation was based on enumeration of 50–70 cells per case. Using a less restrictive cut-off of 10% positive cells, 7 cases (16%) were considered either positive or ‘possibly positive’. Among these, there were 6 of 25 (26%) gastric cancer and 1 of 17 (6%) colon cancer cases.
Figure 2.
Scatterplot and histogram showing percentage of cells with non-fused break-apart signals detected by fluorescence in-situ hybridization (FISH) in each of the two cases of colon and gastric adenocarcinoma employed in this study. There was a single case out of the 42 cases (2.3%) that was positive for anaplastic lymphoma kinase (ALK) rearrangement by FISH using the standard criteria of ⩾15% of cells being positive for the break-apart signal. Using a less-restrictive cut-off of 10% positive cells, 7 cases (16%) were considered either positive or possibly positive.
ALK expression detection by IHC
Using an intensity score of 1 or greater to indicate positive ALK expression, we assessed 0 of 42 cases (0%) as positive. The two cases of lung cancer with known ALK-EML4 translocation that served as positive controls both stained positive with a stronger staining intensity obtained with the Leica antibody in one of the two cases.
Discussion
Signet ring cell carcinoma (SRC) usually arises in the glandular epithelium of the gastrointestinal tract. This histologic subtype is reported in up to 26% of gastric cancers [Taghavi et al. 2012] and in about 2% of colorectal carcinomas [Kwon et al. 2014]. In comparison with adenocarcinoma, SRC arising in the stomach or the colorectal site is more frequent in younger patients, in women and more likely to present at a more advanced stage of disease. Coupled with the predilection for gastric SRC to involve the body and distal stomach and for colonic SRC to present as a right-sided colon cancer, we expect that this subtype of cancer will manifest a distinct biology to match its unique anatomy and histology. Based on the experience in lung cancer where ALK rearranged tumors were more likely to have signet ring features [Shaw et al. 2009], we decided to use tumor histology as an enrichment strategy to facilitate the detection of this subtype of cancer in the expectation that our finding will inform targeted therapeutic strategy leading to a new option for managing SRC of the gastrointestinal tract.
The presence of rearranged ALK gene using IHC or break-apart FISH was required prior to enrollment into the early phase I trial of crizotinib (A8081005). The test was performed at various commercial laboratories prior to the accelerated approval of crizotinib in August 2011. However, FDA approval of crizotinib was based on the presence of ALK translocation in at least 15% of scored cells using FISH as the standard companion diagnostic test [Monjour et al. 1988]. The ALK signaling pathway may be dysregulated due to various mechanisms (activating mutations, gene amplification, translocations). However, the standard FISH test detects only gene rearrangements regardless of the specific fusion companion or breakpoint variant. This could lead to false-negative results if there are complex rearrangements not detectable by the selected FISH probe. Indeed, there was no correlation between maximal tumor reduction in patients treated with crizotinib and the proportion of cells harboring the classic gene alteration within the tumor sample [Camidge et al. 2012]. This well-described limitation of the FISH assay [Weickhardt et al. 2013] could have contributed to the low frequency of ALK positive cases in our series using the previously established cutoff point of 15% from lung cancer patients. The biological significance of a different cut-point of 10% could not be assessed in this retrospective dataset.
We employed IHC as an alternative detection platform to address this limitation of FISH assay since it can detect increased ALK expression that result from different mechanisms of genetic alteration including activating mutations, amplification and translocation [Yi et al. 2011]. With a reported sensitivity comparable with FISH and specificity approaching 100%, IHC is a viable and possibly superior alternative to FISH but requires rigorous tissue processing protocols [Mino-Kenudson et al. 2010]. We failed to detect increased ALK expression by IHC in any of the 42 cases tested in this study, including the single case that was positive by FISH. The failure to show ALK overexpression in the FISH positive case could be due to the discohesive nature of signet ring cancers leading to relatively scanty amount of cellular material against a broad infiltrative stroma [Borger et al. 2007].
Our study used two different platforms (performed in a blinded manner), namely IHC and FISH to carefully study the incidence of ALK gene alterations in gastric and colon SRCs. These complementary techniques demonstrated that ALK gene alteration is a very rare genetic event in SRC. Nonetheless, the identification of a single case both by FISH indicates the potential role of this genetic driver in a small subset of SRC. Our results have further extended the finding by other workers showing a very low frequency of ALK gene alteration in colon cancer. Our detection rate of 4.7% by FISH is comparable to the rate reported by other investigators ranging between 3% and 5% in various gastrointestinal cancer [Aisner et al. 2014]. An enrichment strategy focusing only on SRC histology did not increase the detection rate. Moreover, our inability on IHC to observe increased ALK expression in the single case detected by FISH suggests that caution should be exercised when employing IHC as a surrogate standalone platform for ALK translocation testing, at least in SRC.
In conclusion, ALK gene rearrangement in gastrointestinal cancers is very rare and enrichment strategy focusing on signet ring cell histology did not significantly improve the detection rate. The failure to detect higher number of cases even with the use of different testing platforms further supports the previous work that reported rare occurrence of ALK translocation in gastrointestinal cancers. Further studies are needed to identify prognostic and therapeutic molecular targets in SRC of gastric and colorectal cancers.
Footnotes
Funding: This study was supported by National Institutes of Health (NCI grant 5K23CA164015 to TKO) and Cancer Center Support (grant numbers P30CA138292 and P30-CA46934). BFE, GS, SSR, FRK and TKO are Georgia Cancer Coalition Distinguished Cancer Scholars; OBA is a recipient of NIH T32 CA160040-02 (P.I., Dong M. Shin). The sponsors played no role in the study design, collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Conflict of interest statement: The authors declare no conflict of interest in publishing this article.
Contributor Information
Olatunji B. Alese, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA
Bassel F. El-Rayes, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA
Gabriel Sica, Department of Pathology, Emory University School of Medicine, Atlanta, GA, USA.
Guojing Zhang, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA.
Dianne Alexis, Cancer Tissue and Pathology Shared Resource (CTPSR), Winship Cancer Center of Emory University, Atlanta, GA, USA.
Francisco G. La Rosa, Department of Pathology, University of Colorado School of Medicine, Denver, CO, USA
Marileila Varella-Garcia, Department of Pathology, University of Colorado School of Medicine, Denver, CO, USA.
Zhengjia Chen, Department of Statistics, Rollins School of Public Health, Emory University, Atlanta, GA, USA.
Michael R. Rossi, Department of Radiation Oncology, Emory University School of Medicine, Atlanta, GA, USA
Nazim V. Adsay, Department of Pathology, Emory University School of Medicine, Atlanta, GA, USA
Fadlo R. Khuri, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA
Taofeek K. Owonikoko, Department of Hematology and Medical Oncology, Emory University School of Medicine, 1365 Clifton Road, NE, Room C3080, Atlanta, GA 30322, USA
References
- Aisner D., Nguyen T., Paskulin D., Le A., Haney J., Schulte N., et al. (2014) ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavi P., Jehan Z., Bu R., Prabhakaran S., Al-Sanea N., Al-Dayel F., et al. (2013) ALK gene amplification is associated with poor prognosis in colorectal carcinoma. Br J Cancer 109: 2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger M., Gosens M., Jeuken J., Van Kempen L., Van De Velde C., Van Krieken J., et al. (2007) Signet ring cell differentiation in mucinous colorectal carcinoma. J Pathol 212: 278–286. [DOI] [PubMed] [Google Scholar]
- Camidge D., Theodoro M., Maxson D., Skokan M., O’Brien T., Lu X., et al. (2012) Correlations between the percentage of tumor cells showing an anaplastic lymphoma kinase (ALK) gene rearrangement, ALK signal copy number, and response to crizotinib therapy in ALK fluorescence in situ hybridization-positive nonsmall cell lung cancer. Cancer 118: 4486–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Shin H., Bray F., Forman D., Mathers C., Parkin D. (2010) Estimates of worldwide burden of cancer in 2008: Globocan 2008. Int J Cancer 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- Galetta D., Rossi A., Pisconti S., Colucci G. (2012) The emerging role of ALK inhibitors in the treatment of advanced non-small cell lung cancer. Expert Opin Ther Targets 16(Suppl. 2): S45–S54. [DOI] [PubMed] [Google Scholar]
- Grob T., Heilenkotter U., Geist S., Paluchowski P., Wilke C., Jaenicke F., et al. (2012) Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat 134: 561–567. [DOI] [PubMed] [Google Scholar]
- Jokoji R., Yamasaki T., Minami S., Komuta K., Sakamaki Y., Takeuchi K., et al. (2010) Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol 63: 1066–1070. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S., Woodward W., Yang W., Reuben J., Tepperberg J., Ogura D., et al. (2013) Status of the anaplastic lymphoma kinase (ALK) gene in inflammatory breast carcinoma. Springerplus 2: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E., Bang Y., Camidge D., Shaw A., Solomon B., Maki R., et al. (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K., Shim K., Song E., Choi J., Kim S., Jung H., et al. (2014) Clinicopathological characteristics and prognosis of signet ring cell carcinoma of the stomach. Gastric Cancer 17: 43–53. [DOI] [PubMed] [Google Scholar]
- La Thangue N., Kerr D. (2011) Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat Rev Clin Oncol 8: 587–596. [DOI] [PubMed] [Google Scholar]
- Lin E., Li L., Guan Y., Soriano R., Rivers C., Mohan S., et al. (2009) Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res 7: 1466–1476. [DOI] [PubMed] [Google Scholar]
- Mino-Kenudson M., Chirieac L., Law K., Hornick J., Lindeman N., Mark E., et al. (2010) A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 16: 1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monjour L., Vouldoukis I., Ogunkolade B., Hetzel C., Ichen M., Frommel D. (1988) Vaccination and treatment trials against murine leishmaniasis with semi-purified leishmania antigens. Trans R Soc Trop Med Hyg 82: 412–415. [DOI] [PubMed] [Google Scholar]
- Mosse Y., Wood A., Maris J. (2009) Inhibition of ALK signaling for cancer therapy. Clin Cancer Res 15: 5609–5614. [DOI] [PubMed] [Google Scholar]
- Niu H., Zhou Q., Wang F., Shao Q., Guan Y., Wen X., et al. (2013) Identification of anaplastic lymphoma kinase break points and oncogenic mutation profiles in acral/mucosal melanomas. Pigment Cell Melanoma Res 26: 646–653. [DOI] [PubMed] [Google Scholar]
- Ou S., Kwak E., Siwak-Tapp C., Dy J., Bergethon K., Clark J., et al. (2011) Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol 6: 942–946. [DOI] [PubMed] [Google Scholar]
- Park J., Kerbel R., Kelloff G., Barrett J., Chabner B., Parkinson D., et al. (2004) Rationale for biomarkers and surrogate end points in mechanism-driven oncology drug development. Clin Cancer Res 10: 3885–3896. [DOI] [PubMed] [Google Scholar]
- Popat S., Gonzalez D., Min T., Swansbury J., Dainton M., Croud J., et al. (2012) ALK translocation is associated with ALK immunoreactivity and extensive signet-ring morphology in primary lung adenocarcinoma. Lung Cancer 75: 300–305. [DOI] [PubMed] [Google Scholar]
- Rodig S., Mino-Kenudson M., Dacic S., Yeap B., Shaw A., Barletta J., et al. (2009) Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 15: 5216–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr (2013) Anaplastic lymphoma kinase (ALK): structure, oncogenic activation, and pharmacological inhibition. Pharmacol Res 68: 68–94. [DOI] [PubMed] [Google Scholar]
- Shaw A., Yeap B., Mino-Kenudson M., Digumarthy S., Costa D., Heist R., et al. (2009) Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27: 4247–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M., Choi Y., Enomoto M., Takada S., Yamashita Y., Ishikawa S., et al. (2007) Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- Taghavi S., Jayarajan S.N., Davey A., Willis A.I. (2012) Prognostic Significance of Signet Ring Gastric Cancer. J Clin Oncol 30: 3493–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujii M., Kawano S., Dubois R.N. (1997) Cyclooxygenase-2 expression in human colon cancer cells increases metastatic potential. Proc Natl Acad Sci U S A 94: 3336–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickhardt A., Aisner D., Franklin W., Varella-Garcia M., Doebele R., Camidge D. (2013) Diagnostic assays for identification of anaplastic lymphoma kinase-positive non-small cell lung cancer. Cancer 119: 1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi E., Boland J., Maleszewski J., Roden A., Oliveira A., Aubry M., et al. (2011) Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC Score Algorithm for FISH. J Thorac Oncol 6: 459–465. [DOI] [PubMed] [Google Scholar]