FIG 5.
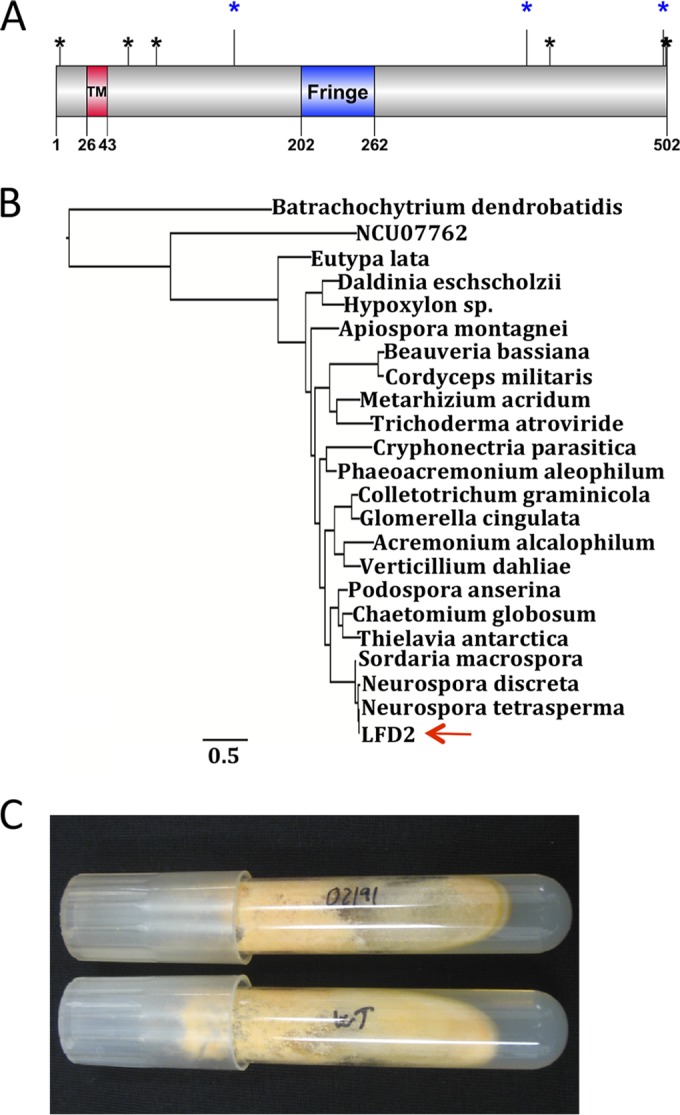
Structure, phylogenetic, and phenotypic analyses of LFD-2. (A) Cartoon of domain structure of LFD-2. The red box shows the transmembrane domain, and the blue box shows the Fringe domain. The blue asterisks indicate predicted N-linked glycosylation sites, and black asterisks indicate predicted O-glycosylation sites (see Materials and Methods). The cartoon was made by DOG (68). (B) Phylogenetic tree for LFD-2 (NCU02191) in the genomes of species from Sordariomycetes: Acremonium alcalophilum (Acral1_c3a.estExt_fgenesh2_pg.C_50864), Apiospora montagnei (e_gw1.92.82.1), Beauveria bassiana (BBA_03605m.01), Chaetomium globosum (CHGT_02214), Colletotrichum graminicola (GLRG_06237T0), Cordyceps militaris (CCM_08528m.01), Cryphonectria parasitica (Crypa1.estExt_Genewise1Plus.C_20699), Daldinia eschscholzii (MIX4991_8_43), Eutypa lata (UCREL1_7331m.01), Glomerella cingulata (e_gw1.9.114.1), Hypoxylon sp. strain CI-4A (gm1.9432_g), Metarhizium acridum (MAC_03592m.01), Neurospora discreta (estExt_Genewise1Plus.C_90445), Neurospora tetrasperma (e_gw1.5.373.1), Phaeoacremonium aleophilum (UCRPA7_6658m.01), Podospora anserina (Pa_5_2960), Thielavia antarctica (e_gw1.19.390.1), Trichoderma atroviride (Triat1.e_gw1.1.1843.1), Verticillium dahliae (VDAG_07638T0), and Sordaria macrospora (SMAC_03630). The LFD-2 homologous sequence in Batrachochytrium dendrobatidis (GP3.005680) and the putative paralog of LFD-2 in N. crassa (NCU07762) were used as outgroups. Arrow points at LFD-2 (NCU02191) from N. crassa. The scale bar indicates the number of substitutions per amino acid. (C) Wild-type culture (bottom) and Δlfd-2 culture (upper) growing on VMM agar.
