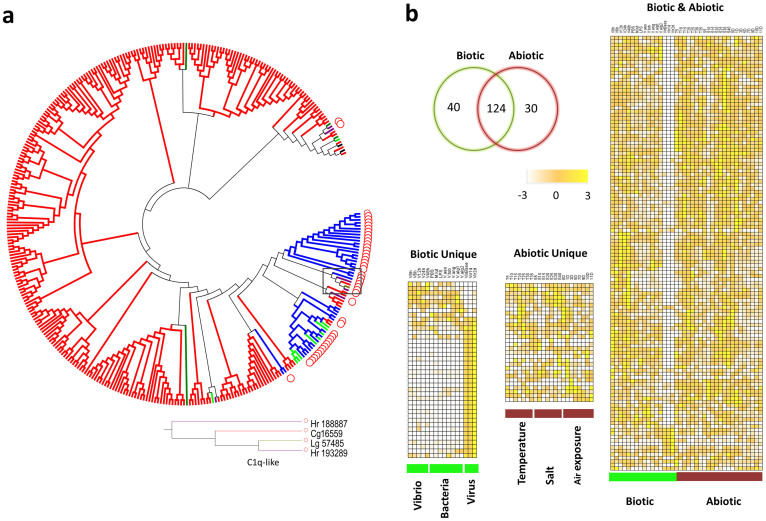
Figure 4. Expansion of C1qDC and divergent expression patterns under challenges and among different organs.
(a), Phylogenetic tree (constructed with the maximum likelihood method) demonstrating lineage-specific expansion of C1qDC in Pacific oyster (Crassostrea gigas – red - Cg) and relationships to: owl limpet (Lottia gigantea – brown - Lg), leech (Helobdella robusta – purple - Hr), sea urchin (Strongylocentrotus purpuratus - green), amphioxus (Branchiostoma floridae - blue) and hydra (Hydra magnipapillata - black) C1qDC sequences. C1qDCs encoding an additional cysteine-rich domain (CRD) domain are noted with red circles. A single C1q-like gene with a collagen domain has been identified in oyster and its phylogenetic tree is shown (above). Scaled expression values are color-coded according to the legend on the top. (b), The number of differentially expressed C1qDCs associated with exposure to biotic and abiotic stressors. Biotic challenges include several species of Vibrio and other bacteria (see Supplementary Table S2 online); abiotic stressors are temperature, salinity and air exposure (see Supplementary Table S2 online). A number of immune genes function in abiotic stress. Heat maps from left to right are expression patterns of C1qDC: only under biotic challenges, only under abiotic challenges and those C1qDCs up-regulated by both biotic and abiotic challenges.