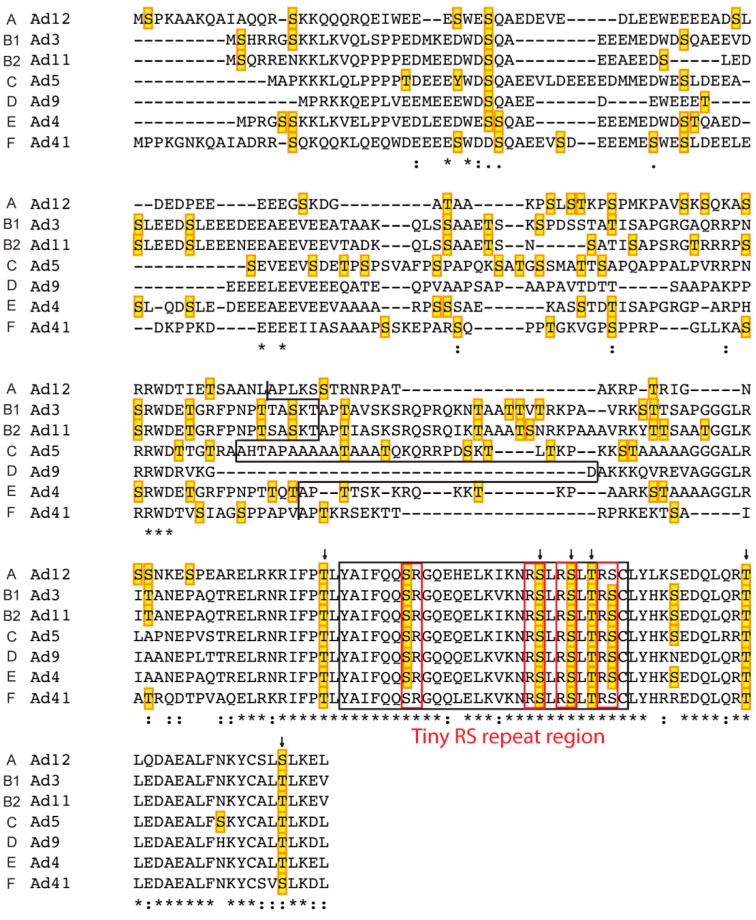
Figure 5.
Alignment of L4-33K proteins from different human serotypes and their putative phosphorylation sites. The amino acid sequence of the indicated L4-33K proteins was aligned using Clustal Omega software. The conserved residues are indicated by star (*), colon (:), and period (.) according to the level of identity and similarity of the amino acid aligned: full identity, amino acids with similar characteristics, and weakly similar residues, respectively. The black line separates the N-terminus (shared with the respective L4-22K) and the C-terminal region of each serotype. The putative phosphorylation sites detected with NetPhosK 1.0 are marked in yellow boxes, and the conserved residues predicted to be phosphorylated by the same kinase are indicated by arrows (see also Table S1B). The ds domain is shown with a black-box with the tiny SR repeats indicated in red boxes.