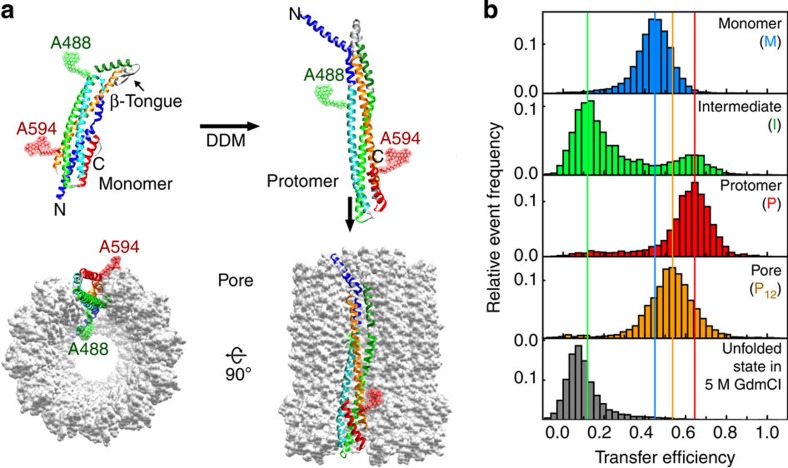
Figure 1. The different conformational states of labelled ClyA on mixing with DDM as observed by single-molecule FRET.
(a) Crystal structures of the monomer and the ClyA protomer in the context of the pore complex (PDB code 1QOY18 and PDB code 2WCD17). The protomer conformation is represented by one pore subunit. N and C indicate the N- and carboxy termini of the protein, and A488 and A594 the positions labelled with Alexa Fluor 488 (position 56) and 594 (position 252), respectively. Structure representations were created with Chimera63 and Avogadro64, colour scheme according to Mueller et al.17 (b) Transfer efficiency histograms of the different ClyA species that can be distinguished by single-molecule FRET. The histograms (each containing ≳ 5,000 events) were normalized to an area of 1. The coloured vertical lines indicate the peak positions of the species in the transfer efficiency histograms. The histogram of the monomer (M) was recorded in the absence of DDM. The histograms of intermediate (I) and protomer (P) are from the kinetic measurements in the presence of DDM at a ClyA concentration of ~100 pM at 55 and 1,765 s, respectively, after mixing with DDM. For the histogram of the pore (P12), 5 μM ClyA with 1% labelled ClyA were incubated with DDM for 2 h and the formed pores were purified by size-exclusion chromatography according to Eifler et al.7 The resulting transfer efficiency is identical within uncertainty to what we observe in single-molecule kinetic measurements at times where oligomer formation is complete (Fig. 4a and Supplementary Fig. 8a). The difference between the transfer efficiencies of the oligomer state and the protomer can largely be assigned to acceptor dye quenching in the complex, rather than a difference in conformation (see Supplementary Table 1).