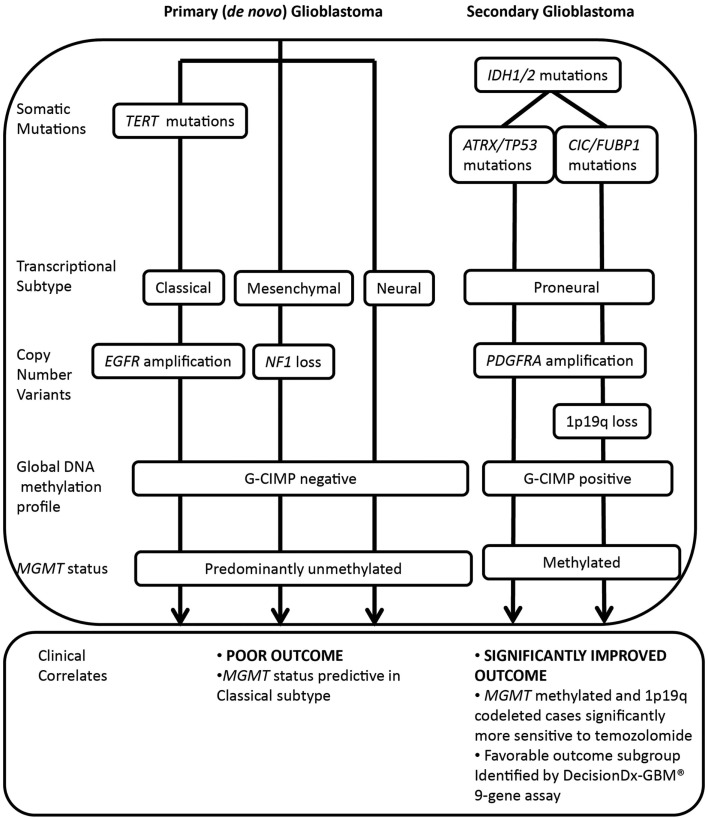
Figure 1.
Molecular classification of major glioblastoma subtypes and correlation with treatment response and outcome. IDH1/2 mutations are major prognostic biomarkers, stratifying primary and secondary pathways of gliomagenesis. Primary glioblastoma features a high frequency of TERT mutations, whereas IDH1/2 mutated glioma (including secondary glioblastoma and low grade glioma) may be further subdivided on the basis of co-mutations in either ATRX and TP53 or CIC and FUBP1, occurring at high frequency in astrocytic or oligodendroglial tumor subtypes. Co-deletion of 1p and 19q, a marker of enhanced chemosensitivity, also clusters with mutations in CIC and FUBP1. Primary glioblastomas display classical, mesenchymal, and neural phenotypes, whereas secondary glioblastomas tend to display a proneural phenotype that shifts toward a mesenchymal phenotype with recurrence. The significantly improved outcome of the proneural subset is due to the G-CIMP phenotype, established by IDH1/2-mediated metabolic reprogramming of the epigenome. Primary glioblastomas and a subset of proneural tumors are glioma-CpG island hypermethylator phenotype (G-CIMP) negative, and a large proportion are MGMT unmethylated. Both classical and mesenchymal transcriptional subtypes benefit from concurrent chemoradiotherapy, however, MGMT status is only predictive of treatment response in the classical subset.