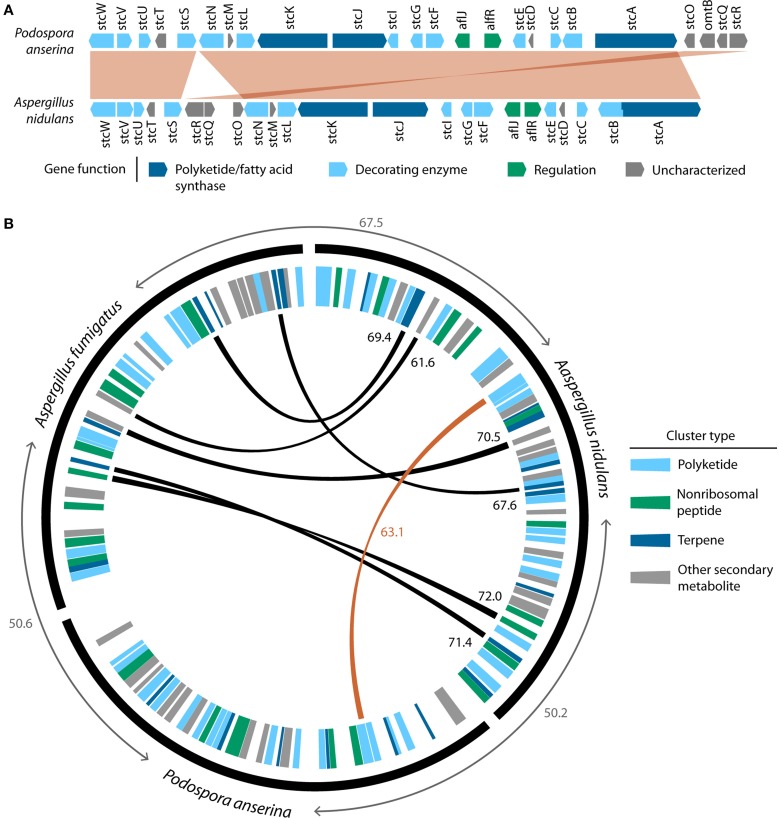
Figure 2.
The remarkable similarity of the sterigmatocystin MGC between Aspergillus nidulans and Podospora anserina, two organisms belonging to different fungal classes, is evidence of HGT. (A) Synteny conservation between the sterigmatocystin MGC from Podospora anserina and Aspergillus nidulans. Aligned regions were drawn using the progressive Mauve algorithm (Darling et al., 2010) and are shown in red. (B) Conservation of SM gene clusters between Podospora anserina, Aspergillus nidulans and Aspergillus fumigatus. Circular tracks were created using Circos (http://circos.ca); the outer black track shows the relative gene counts in each of the three species. However, to visualize the relative location of SM gene clusters, the width of genes in these clusters have been drawn at 20 times the width of unclustered genes. Average amino acid percent identities (%IDs) of all reciprocal best BLAST hits (RBBHs) between the three genomes are shown in gray. SM gene clusters were predicted using antiSMASH (Blin et al., 2013), and SM cluster type is indicated by the colored wedges of the inner track. SM clusters were considered homologous if greater than 50% of their genes were RBBHs. Black links and black numbers indicate homologous SM clusters between Aspergillus nidulans and Aspergillus fumigatus and the average %IDs of RBBHs of the clustered genes, respectively. The red link and red number indicate the only homologous SM cluster (sterigmatocystin) identified between Aspergillus nidulans and Podospora anserina and the average %ID of RBBHs of the clustered genes, respectively. There are no homologous SM clusters between Aspergillus fumigatus and Podospora anserina.