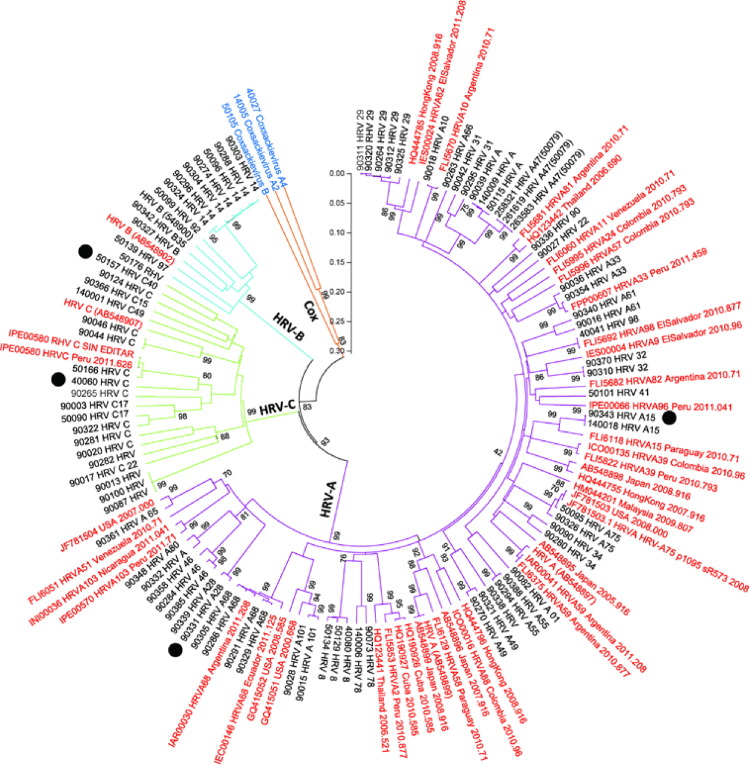
Fig. 2.
Phylogenetic tree of partial HRV VP4/VP2 RNA sequences from 84 study samples. Prototype strains of HRV-A–C from GenBank were included for comparison (Clustal X version 2.0.1). The tree was constructed using the neighbor-joining method in MEGA software (version 5). The statistical significance of the tree topology was tested by bootstrapping (1000 replicas). Pairwise distances between and within the genotypes at the nucleotide level were calculated with Kimura 2 parameters and with Poisson correction at the amino acid level with MEGA software. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Black dot: hospitalized case, sequence name in red: standard sequence, Cox: coxsackievirus.