Figure 6.
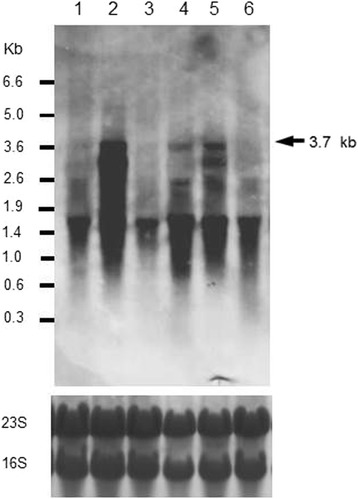
Northern blotting analysis of rocG transcripts in the presence and absence of lctE. RNAs were prepared from 168 (lane 1), TM023 (ccpA::neo, lane 2), MY02 (lctE::spc, lane 3), 1A95 (degU32, lane 4), TM024 (degU32 ccpA::neo, lane 5), and KI004 (degU32 lctE::spc, lane 6) cells for northern blotting analyses using probes for rocG transcripts under high-stringency conditions. Experiments were repeated independently at least three times with similar results, and representative data are shown. At the bottom, rRNAs (23S and 16S) were visualized as loading controls using methylene blue staining.
