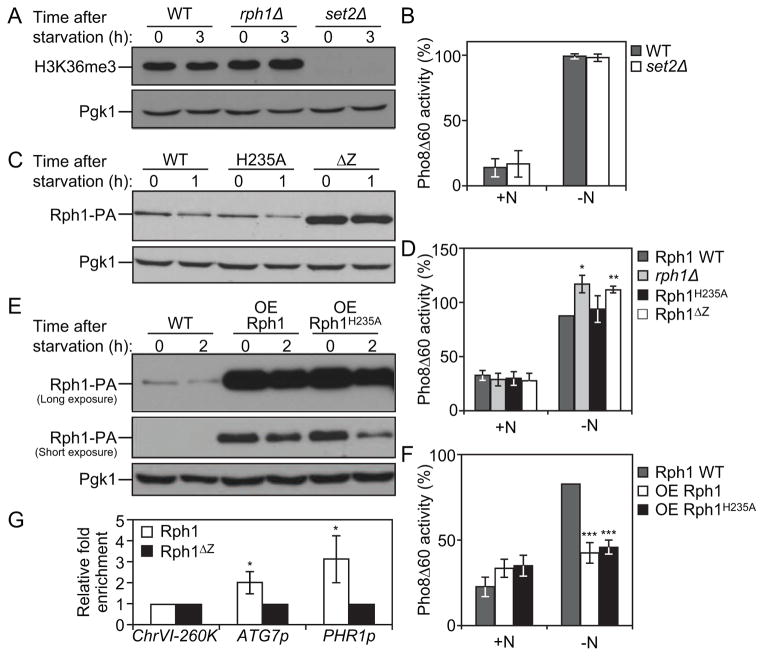
Figure 5. Rph1 DNA binding ability but not histone demethylase activity is required for its function in autophagy.
(A) Wild-type (YTS158), rph1Δ (YAB300) and set2Δ (YAB318) cells were grown in YPD (+N) and then starved for nitrogen (−N) for 3 h. Protein extracts were analyzed by western blot with anti-H3K36me3 antibody and anti-Pgk1 (loading control) antiserum.
(B) Wild-type (YTS158, BY4742) and set2Δ (YAB318) cells were grown in YPD (+N) and then starved for nitrogen (−N) for 3 h. The Pho8Δ60 activity was measured and normalized to the activity of wild-type cells, which was set to 100%. For panels B, D, F and G, the data represent the average of 3 independent experiments ± standard deviation.
(C–D) rph1Δ cells were transformed with the RPH1p-RPH1-PA (Rph1, Wild-type), RPH1p-RPH1H235A-PA (H235A), or RPH1p-RPH1ΔZ-PA (ΔZ) plasmids or the corresponding empty plasmid (pRS461-PA, rph1Δ). Cells were grown in rich selective medium (SMD-Ura) until mid-log phase and then starved for nitrogen for 1 h.
(C) The stability of Rph1 mutants was analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum.
(D) The Pho8Δ60 activity was measured and normalized to the activity of wild-type Rph1 cells after 1 h of nitrogen starvation (−N), which was set to 100%.
(E–F) Rph1-PA cells (WT, YAB366) and cells overexpressing Rph1-PA (OE Rph1, YAB363) or Rph1H235A-PA (OE Rph1H235A, YAB364) were grown in YPD (+N) until mid-log phase and then starved for nitrogen for 2 h (−N).
(E) Protein extracts were analyzed by western blot with either an antibody that recognizes PA or anti-Pgk1 (loading control) antiserum.
(F) The Pho8Δ60 activity was measured and normalized to the activity of wild-type cells after 2 h of nitrogen starvation (−N), which was set to 100%.
(G) Rph1-PA binds the ATG7 promoter. rph1Δ cells transformed with the RPH1p-RPH1-PA (Rph1) or RPH1p-RPH1ΔZ-PA (Rph1ΔZ) plasmids were analyzed by ChIP. ChIP was conducted on the ATG7 promoter (ATG7p), a large non-coding region located at 260 kb on chromosome VI (ChrVI-260K) which was used as a negative control, and on the PHR1 promoter (PHR1p) which was used as a positive control. Results were normalized to the input DNA and calibrated to the ChrVI-260K PCR product; results are presented as fold-enrichment of Rph1 binding compared to Rph1ΔZ, which was set to 1.