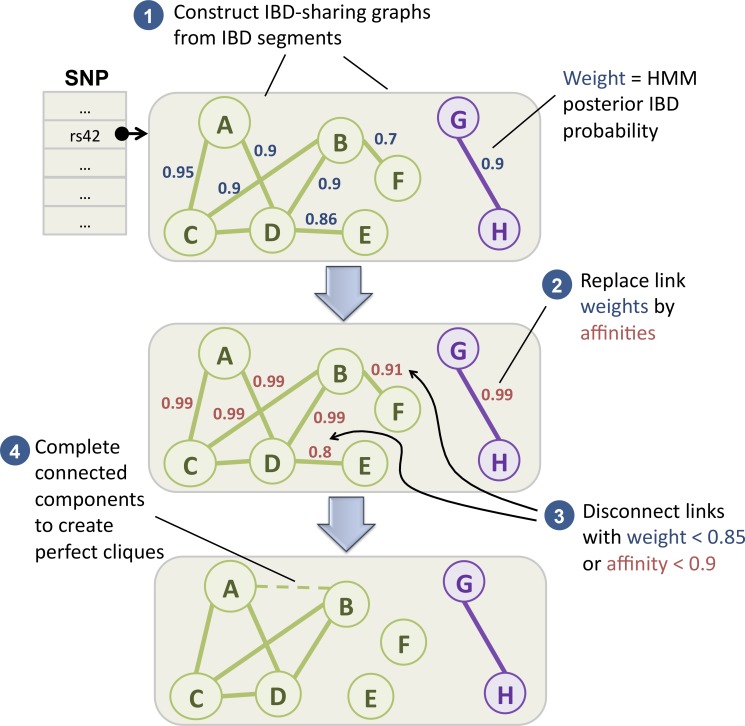
Fig 2. Partitioning an IBD-sharing graph into cliques.
(1) IBD segments are indexed into a graph at each SNV. Nodes represent haplotypes (denoted A-H). Each pair of haplotypes that share an IBD segment at the SNV is connected with a link whose weight equals the HMM posterior probability. (2) Link weights are replaced by affinities. Links with small original weight or affinity are removed (3); all nodes within each of the resulting connected components are connected (4).