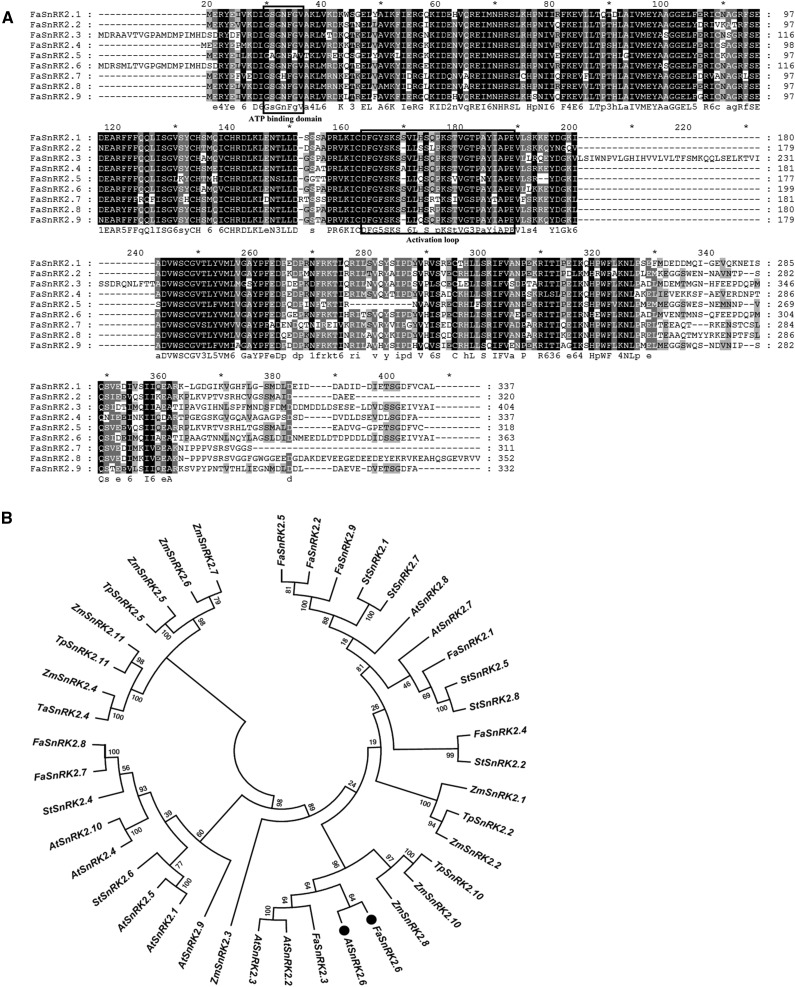
Figure 1.
Phylogenetic analysis of SnRK2.6 proteins. A, Sequence alignments of the deduced amino acid sequences of FaSnRK2s. The deduced amino acid sequences of FaSnRK2.1 to FaSnRK2.9 were aligned using ClustalX 2.0.12 software with default settings. The alignments were edited and marked using GeneDoc. Black and light-gray shading indicate identical and similar amino acid residues, respectively. The predicted functional domains are boxed. B, Phylogenic analysis of SnRK2 homologs from various plant species. The phylogenetic trees were constructed using the neighbor-joining method in MEGA6 software with 1,000 bootstrap replicates. The numbers at nodes represent bootstrap values. OST1 and its ortholog in strawberry are marked with black circles. Species name abbreviations are as follows: At, Arabidopsis; Np, Nicotiana plumbaginifolia; St, potato (Solanum tuberosum); Ta, wheat (Triticum aestivum); Tp, Triticum polonicum; and Zm, maize.