The green algae Chlamydomonas reinhardtii possesses at least two SUMO conjugates, one involved in housekeeping and the other in response to stress.
Abstract
Posttranslational modification of proteins by small ubiquitin-like modifier (SUMO) is required for survival of virtually all eukaryotic organisms. Attachment of SUMO to target proteins is catalyzed by SUMO E2 conjugase. All haploid or diploid eukaryotes studied to date possess a single indispensable SUMO conjugase. We report here the unanticipated isolation of a Chlamydomonas reinhardtii (mutant5 [mut5]). in which the previously identified SUMO conjugase gene C. reinhardtii ubiquitin-conjugating enzyme9 (CrUBC9) is deleted. This surprising mutant is viable and unexpectedly, displays a pattern of protein SUMOylation at 25°C that is essentially identical to wild-type cells. However, unlike wild-type cells, mut5 fails to SUMOylate a large set of proteins in response to multiple stress conditions, a failure that results in a markedly reduced tolerance or complete lack of tolerance to these stresses. Restoration of expected stress-induced protein SUMOylation patterns as well as normal stress tolerance phenotypes in mut5 cells complemented with a CrUBC9 gene shows that CrUBC9 is an authentic SUMO conjugase and, more importantly, that SUMOylation is essential for cell survival under stress conditions. The presence of bona fide SUMOylated proteins in the mut5 mutant at 25°C can only be explained by the presence of at least one additional SUMO conjugase in C. reinhardtii, a conjugase tentatively identified as CrUBC3. Together, these results suggest that, unlike all other nonpolyploid eukaryotes, there are at least two distinct and functional SUMO E2 conjugases in C. reinhardtii, with a clear division of labor between the two sets: One (CrUBC9) is involved in essential stress-induced SUMOylations, and one (CrUBC3) is involved in housekeeping SUMOylations.
The modification of proteins by small ubiquitin-like modifier (SUMO) in eukaryotes is considered essential for normal cell growth and development (Geiss-Friedlander and Melchior, 2007). Posttranslational modification of a protein by SUMO occurs in an enzymatic pathway highly analogous to the ubiquitination pathway and involves three types of enzymes: a SUMO E1 activase, a SUMO E2 conjugase, and a SUMO E3 ligase (Geiss-Friedlander and Melchior, 2007). The role of the SUMO E2 conjugase is to catalyze the formation of an isopeptide bond that connects the C terminus of SUMO to the ε-amino group of a Lys residue within a target protein (Matunis et al., 1996; Desterro et al., 1997; Johnson and Blobel, 1997). Across a wide range of phylogenies of haploid and diploid organisms, a single gene has been identified that encodes for a SUMO E2 conjugase. SUMO E2 conjugase mutations that cause loss of function are lethal, strongly suggesting that SUMOylation is essential for cell viability (Seufert et al., 1995; Hayashi et al., 2002; Nacerddine et al., 2005; Saracco et al., 2007). In the fission yeast Schizosaccharomyces pombe, deletion of the hydroxy urea sensitive5 (HUS5) SUMO E2 conjugase gene results in an hus5 mutant that, although viable, shows severe growth defects (al-Khodairy et al., 1995), which is, again, consistent with an essential role for SUMOylation in normal cell growth. SUMO E2 conjugases are similar in sequence to ubiquitin conjugase enzymes, and initial reports of various SUMO E2 conjugase genes before the identification of SUMO as a posttranslational modification led to their prediction as ubiquitin-conjugating enzymes (UBCs; al-Khodairy et al., 1995; Seufert et al., 1995). However, although there is analogy or even homology between the given components of the two pathways, the SUMOylation and ubiquitination cascades are nonoverlapping. The SUMO E2 conjugase cannot form a thioester with ubiquitin, and similarly, ubiquitin conjugases are incapable of forming thioesters with SUMO (Desterro et al., 1997).
Another important distinction between the ubiquitin and SUMO pathways is the number of genes identified for each component in the pathway. Given the vast number of proteins modified by SUMO or ubiquitin, proper selection of targets, timing of modification, and intracellular localization of these reactions require a great deal of regulation and specificity. The ubiquitin pathway accomplishes this through the use of numerous E2 conjugases and E3 ligases in various combinations. As an example, as many as 25 ubiquitin E2 conjugases have been identified in the Arabidopsis (Arabidopsis thaliana) genome along with hundreds of potential ubiquitin E3 ligases (Bachmair et al., 2001; Kraft et al., 2005). Compared with the hundreds of putative ubiquitin E3 ligases in Arabidopsis, only two SUMO E3 ligases have been identified (Miura et al., 2005; Huang et al., 2009; Ishida et al., 2009). In striking contrast to the numerous ubiquitin E2 conjugase enzymes identified in Arabidopsis and other eukaryotes, only a single SUMO E2 enzyme has been identified in virtually every organism for which the SUMOylation pathway has been characterized (Seufert et al., 1995; Yasugi and Howley, 1996; Hayashi et al., 2002; Nacerddine et al., 2005; Saracco et al., 2007). A technical exception to this rule is provided by several land plants that have undergone various rounds of polyploidization or entire genome fusions during interspecies hybridizations (Renny-Byfield and Wendel, 2014) and, thus, contain multiple SUMO E2 conjugase genes (Novatchkova et al., 2012). Two other technical exceptions are zebrafish (Dana rerio; Nowak and Hammerschmidt, 2006) and rice (Oryza sativa; Nigam et al., 2008), which contain two nearly identical SUMO E2 conjugase genes that are apparent products of recent gene duplications.
Thus, there exists a marked dichotomy between the need of cells for multiple ubiquitin E2 conjugases to posttranslationally modify proteins in response to diverse external and internal signals and the ability of a single SUMO E2 conjugase to accomplish all cellular SUMOylation reactions in response to the same diversity of signals (Geiss-Friedlander and Melchior, 2007).
In addition to the requirement for protein SUMOylation to maintain cell viability, changes in SUMOylation patterns have long been associated with response to stress conditions. In mammalian cells, SUMO1 is predominantly conjugated to proteins under nonstress conditions, whereas SUMO2 and SUMO3 are conjugated to proteins in response to a variety of abiotic stresses, including heat stress, salt stress, and oxidative stress (Saitoh and Hinchey, 2000; Šramko et al., 2006). Similar observations have been made in regard to the preferential use of the proteins AtSUMO1 and AtSUMO2 for SUMOylation in response to a variety of stresses in Arabidopsis (Kurepa et al., 2003; Saracco et al., 2007). The types of proteins modified by SUMO in several different organisms under stress conditions have been identified through proteomic analysis and typically include several hundred different proteins (Golebiowski et al., 2009; Miller et al., 2010; Bruderer et al., 2011; Miller and Vierstra, 2011). Given that SUMO E2 conjugase deletion mutants are lethal, the importance of SUMOylation in stress response has been determined largely from analyzing viable individual E3 ligase and SUMO protease mutants in response to stress. Mutants of the Arabidopsis E3 ligase SIZ1 show increased sensitivity to phosphate deprivation, excess copper, temperature stress, and drought stress (Miura et al., 2005, 2007; Yoo et al., 2006; Catala et al., 2007; Chen et al., 2011). Interestingly, siz1 (E3 ligase) mutants in Arabidopsis show increased tolerance to salt stress (Miura et al., 2011), whereas the SUMO protease (isopeptidase) double mutant, ots1 ots2 (for overly sensitive to salt1 overly sensitive to salt2) shows extreme sensitivity to salt (Conti et al., 2008). Another indirect indication of the role of SUMOylation in the response of plants to stress is the recent observation linking changing levels of SUMOylation to changing levels of salicylic acid (Villajuana-Bonequi et al., 2014).
The role of SUMO during stress is best understood in response to heat stress. The heat shock-associated transcription factors (HSFs) HSF1, HSF2, and HSF4b are modified by SUMO in mammalian cells (Goodson et al., 2001; Hong et al., 2001; Hietakangas et al., 2006). SUMOylation increases the DNA binding activity of these transcription factors (Goodson et al., 2001; Hong et al., 2001). In addition, HSF1, which modulates the induction of heat shock protein gene expression in response to elevated temperatures, is SUMOylated upon heat stress, an event that prompts relocalization of HSF1 to nuclear granules (Hong et al., 2001).
In Arabidopsis, HSFA2 is targeted for SUMOylation during extended exposure to heat stress and recovery (Cohen-Peer et al., 2010). In this case, SUMOylation negatively regulates the activity of this transcription factor. In addition to heat stress, the SUMOylation system of plants plays a key role in response to numerous other biotic and abiotic stress conditions (Šramko et al., 2006; Park and Yun, 2013; Xu and Yang, 2013).
In our initial characterization of the SUMOylation system of Chlamydomonas reinhardtii, we identified a protein named CrUBC9 as the most likely SUMO E2 conjugase in this organism and showed that it possessed SUMO E2 conjugase activity in vitro (Wang et al., 2008). Additionally, it was shown by immunoblot analysis that SUMOylation patterns markedly changed in response to various abiotic stress conditions (Wang et al., 2008). Because in virtually every other haploid or diploid organism studied, a single SUMO E2 conjugase had been identified, it was presumed that CrUBC9 was likely the only SUMO E2 conjugase present in the C. reinhardtii genome and that it was essential for cell viability. However, here, we report the isolation of a viable deletion mutant, mut5, which lacks the CrUBC9 gene and yet, displays no obvious growth defect under nonstress conditions. The viability of mut5 allowed a unique opportunity to study the role of the SUMOylation changes previously observed under abiotic stress conditions. In addition, if CrUBC9 was the only functional SUMO conjugase in C. reinhardtii, the viability of mut5 presented an intriguing set of possibilities: (1) C. reinhardtii does not require SUMOylation for viability (because mut5 was clearly viable and apparently healthy), or (2) C. reinhardtii contains at least two functional SUMO conjugases. We report here that CrUBC9 is essential for SUMOylation in response to abiotic stress and that this SUMOylation response is required for cell growth and/or survival during exposure to stress conditions. We also document the presence of SUMOylated proteins in mut5 under nonstress conditions, despite the complete lack of CrUBC9, indicating the presence of an additional functional SUMO conjugase. Bioinformatic analysis is presented indicating that CrUBC3 is likely the second SUMO conjugase. Together, these data provide strong evidence that C. reinhardtii is unique among other organisms in that it contains two distinct SUMO E2 conjugases with a distinct division of labor in carrying out SUMOylation functions (i.e. a stress-specific SUMO conjugase, CrUBC9, that is indispensable for SUMOylation and cell survival under stress conditions and a second SUMO E2 conjugase, CrUBC3, that is responsible for SUMOylations under nonstress conditions.
RESULTS
mut5 Contains a Complete Deletion of the CrUBC9 Gene
In the course of screening for mutants defective in RNA interference (Ibrahim et al., 2006, 2010), the mutant, mut5, was identified. Although mut5 lacked a strong RNA interference phenotype, sequencing of DNA in proximity to the insertion site of the paromomycin resistance selectable marker gene used to generate mut5 revealed a deletion of nucleotides 9166813 to 9188745 on chromosome 2 of the C. reinhardtii genome (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). This deletion includes the entire CrUBC9 gene (Protein ID 57440) as well as four neighboring genes, including three nonannotated genes (Protein IDs 158096, 148626, and 148627) and the predicted protein Motile19 (Protein ID 173947). Despite deletion of the presumably essential CrUBC9 gene, mut5 was viable and grew at normal rates when cultured at 25°C (see data below).
CrUBC9 Is Dispensable for SUMOylation under Nonstress Conditions But Required for Stress-Induced SUMOylation
It was previously shown that multiple high-Mr proteins are SUMOylated in C. reinhardtii in response to a heat shock of 42°C (Wang et al., 2008). To determine if mut5, which lacks the CrUbc9 gene, is capable of this stress-induced SUMOylation, wild-type and mut5 cultures were shifted to 42°C for 1 h, and the patterns of SUMOylation in these cultures were detected and analyzed on immunoblots using anti-SUMO antibodies (Fig. 1). Wild-type cells grown at 25°C show one major protein at approximately 130 kD but few, if any, other high-Mr SUMOylated proteins. In comparison, wild-type cells exposed to 42°C contain many high-Mr proteins (>65 kD) modified by SUMO as previously observed. These newly modified proteins are detectable within 30 min after the shift of cells to 42°C and persist throughout prolonged heat stress (Supplemental Fig. S1). Interestingly, mut5 cells grown at 25°C show a similar pattern of SUMOylation compared with wild-type cells grown at 25°C, despite the complete lack of a functional CrUBC9 gene. In marked contrast to wild-type cells, mut5 cells shifted to 42°C lack the ability to support SUMOylation of high-Mr proteins and, instead, show a SUMOylation pattern that appears identical to nonstressed cells (Fig. 1; Supplemental Fig. S1). These results show that mut5 is incapable of SUMOylating proteins in response to a shift to 42°C and therefore, suggest that the CrUBC9 protein is required for heat-stress induced SUMOylation.
Figure 1.
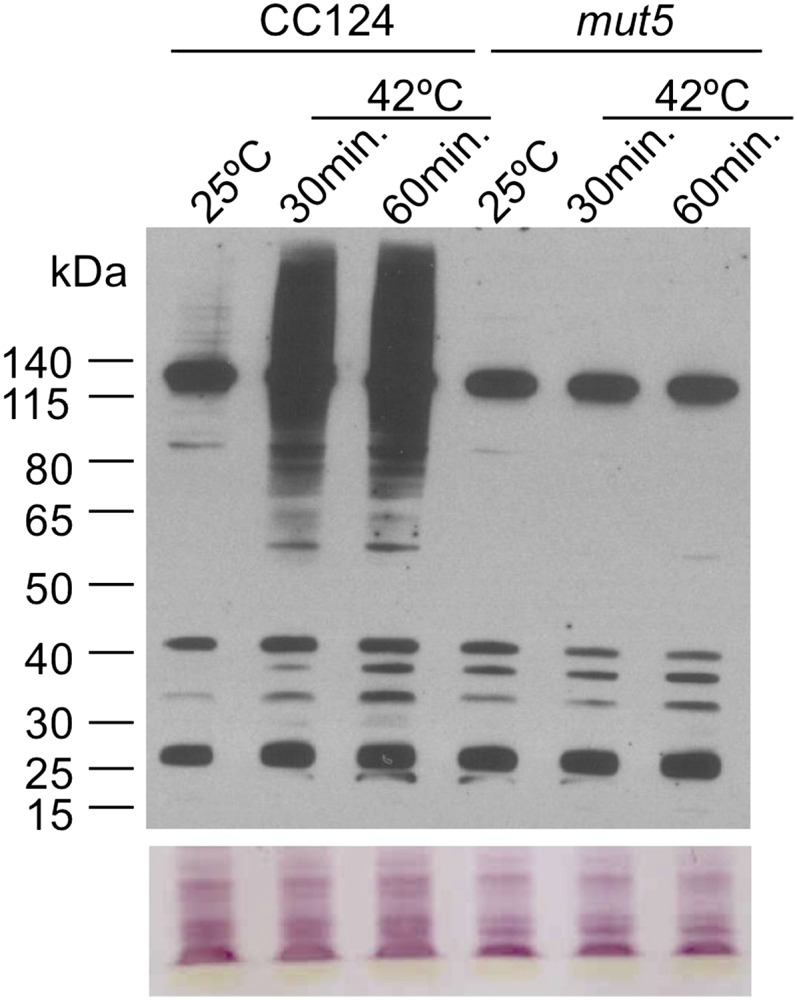
mut5 fails to accumulate high Mr SUMOylated proteins in response to heat shock at 42°C. Protein-blot analysis was performed with total cell extracts of C. reinhardtii CC124 and mut5 cultures grown at 25°C (lanes 1 and 4) or after shifting of these cells to 42°C for 30 or 60 min (lanes 2 and 3, CC124; and lanes 5 and 6, mut5, respectively). SUMOylated proteins in these cell extracts were separated on BisTris SDS-PAGE gels and detected on protein blots with anti-SUMO antibodies (upper). Extracts from equal numbers of cells were loaded into each lane, and staining of the protein blot with Reactive Brown stain was used to show similar loading between lanes (lower).
SUMOylated Proteins Are Present in mut5 Despite the Lack of a Functional CrUBC9 E2 Conjugase
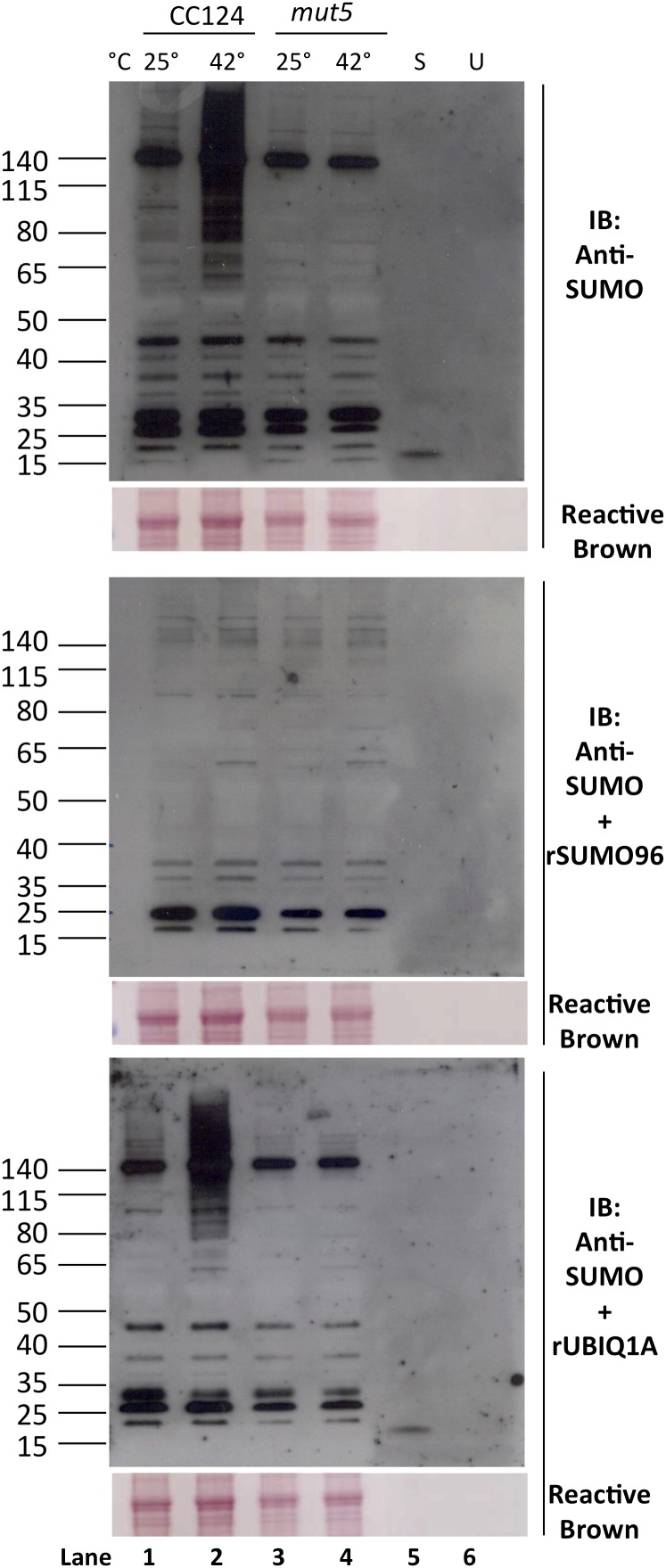
The pattern of proteins detected with anti-SUMO antibodies in mut5 extracts appeared identical to the pattern of proteins detected in nonstressed wild-type cells grown at 25°C (Fig. 1). Because mut5 has a complete deletion of the CrUBC9 gene, the presence of SUMOylated proteins in mut5 strongly suggests the presence of at least one additional functional SUMO E2 conjugase in the C. reinhardtii genome. To confirm that the bands detected in these blots are, indeed, SUMOylated proteins, a competition assay was performed by preincubating anti-SUMO antibodies with excess recombinant SUMO to sequester SUMO antibodies and thus, diminish or ablate the signal obtained from any bona fide SUMO proteins on protein blots of cell extracts. Recombinant CrSUMO96, the protein against which the anti-SUMO antibodies were generated, was preincubated with anti-SUMO antibodies before application to a protein blot of wild-type and mut5 extracts isolated from cells maintained at either 25°C or 42°C. As a negative control, anti-SUMO antibodies were also preincubated with either a nonfat dry milk-based blocking reagent alone or recombinant C. reinhardtii Ubiquitin 1A (CrUBIQ1A). These mixtures were then applied independently to identical protein blots (Fig. 2). In blots of proteins from wild-type cells, recombinant CrSUMO96 competed away the signal of all of the high-Mr proteins (greater than 65 kD) in extracts isolated from cells subjected to 42°C, indicating that these proteins are, indeed, SUMOylated in response to heat shock. In addition, in blots of proteins from both nonstressed wild-type and mut5 cell extracts, signals from several bands were competed away, including the prominent band at approximately 130 kD (Fig. 2, middle). These observations confirm that SUMOylation of proteins occurs in both mut5 and wild-type cells under nonstress conditions in addition to the SUMOylation of new proteins that occurs under stress conditions in wild-type cells. Importantly, preincubation of the anti-SUMO antibodies with recombinant ubiquitin (CrUBIQ1A) failed to compete away the signal from those same proteins, indicating that the antibodies are specific for SUMO (Fig. 2, bottom). Furthermore, the anti-SUMO antibodies failed to recognize recombinant CrUBIQ1A on those same blots (Fig. 2, lane U), suggesting that the anti-SUMO antibodies do not cross react with ubiquitin and are specifically recognizing SUMOylated proteins. Together, these data provide compelling evidence that at least one SUMO conjugase in addition to CrUBC9 is present in C. reinhardtii cells.
Figure 2.
mut5 extracts contain SUMOylated proteins. Proteins in extracts of wild-type and mut5 cells previously incubated at 25°C or for 1 h at 42°C (along with recombinant SUMO96 [S] and UBIQ1A [U]) were separated by electrophoresis on BisTris SDS-PAGE gels and used to prepare three identical protein blots. The immunoblot (IB) shown at top was incubated with anti-SUMO antibodies alone. Protein blots shown in middle and bottom were incubated with anti-SUMO antibodies preincubated with saturating amounts of CrSUMO96 (middle) or CrUBIQ1A (bottom).
CrUBC3 Is Most Likely the Second SUMO E2 Conjugase Gene in C. reinhardtii
Previous analyses of potential SUMO E2 conjugases in C. reinhardtii (Wang et al., 2008) suggested that, among the candidate genes, CrUBC9 was the most similar to verified SUMO E2 conjugase genes found in other eukaryotic organisms. In that same analysis, however, it was noted that the distinction between some of the candidate enzymes as SUMO conjugases versus ubiquitin conjugases was uncertain (Wang et al., 2008). Given that the data presented above confirm the presence of SUMOylated proteins in mut5, we sought to determine which of the other three candidate proteins might be the second SUMO conjugase that is responsible for SUMOylation under nonstress conditions.
To distinguish between SUMO-conjugating enzymes and UBCs, an amino acid sequence alignment was generated that allowed comparisons of the four C. reinhardtii proteins previously identified as potential SUMO conjugase enzymes (including CrUBC9) with the bona fide SUMO-conjugating enzyme from yeast (Saccharomyces cerevisiae; ScUBC9) as well as all verified UBCs from this organism that were similar in length to the candidate conjugase enzymes from C. reinhardtii (Supplemental Fig. S2). The crystal structure of the mouse/human SUMO conjugase identified an insertion of amino acids in the N-terminal region of the protein compared with ubiquitin conjugase enzymes (Tong et al., 1997). The alignment in Supplemental Figure S2 reveals a similar insertion for ScUBC9 compared with the yeast ubiquitin conjugase enzymes as well as just two C. reinhardtii proteins, CrUBC9 and CrUBC3. This insert includes two residues (DG) that are 100% conserved among all three and also present in the 5-amino acid insertion previously identified in human and mouse Ubc9 proteins (Tong et al., 1997). Similarly, a comparison of the sequences reveals that, for both CrUBC9 and CrUBC3, the yeast SUMO conjugase is the best yeast homolog in the alignment (61.78% and 52.60% identity to ScUBC9, respectively), whereas for the other two C. reinhardtii proteins, a yeast ubiquitin conjugase is a better ortholog (Supplemental Table S1).
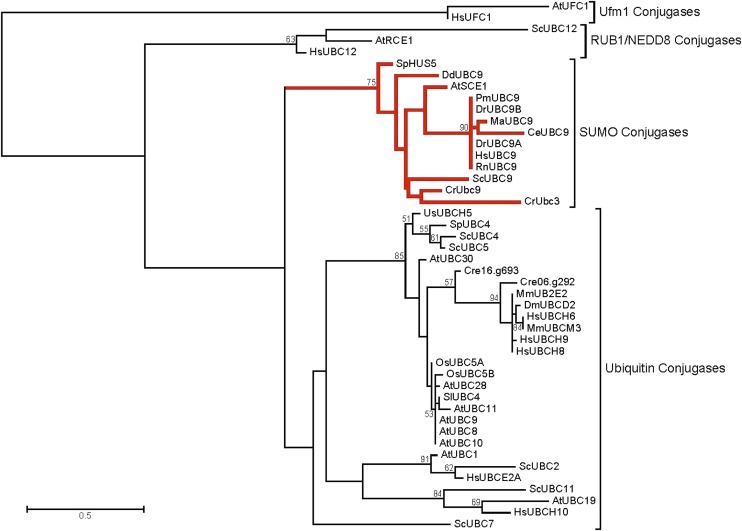
A phylogenetic tree generated using the top BLAST hits from the UniProt/SwissProt database for each of four proteins as well as additional E2 conjugase enzymes revealed that CrUBC9 and CrUBC3 clustered with known SUMO E2 conjugases, whereas the other two previously identified proteins cluster with known ubiquitin E2 conjugases (Fig. 3).
Figure 3.
Phylogenetic tree showing clustering of SUMO and ubiquitin E2 conjugases with their respective conjugases from C. reinhardtii. This map shows the relationship between known and predicted SUMO conjugase and ubiquitin conjugase enzymes as well as candidate SUMO conjugase enzymes from C. reinhardtii. CrUBC3 and CrUBC9 cluster with known SUMO E2 conjugases, whereas the other two C. reinhardtii candidates cluster with verified ubiquitin E2 conjugases. Annotation information for the tree shown is available in Supplemental Table S2.
As an initial comparison of CrUBC9 and CrUBC3, transcript levels for mRNAs encoding these two proteins were analyzed under nonstress (25°C) and stress (42°C) conditions (Supplemental Fig. S3). Under nonstress conditions, CrUBC9 mRNA is more abundant (approximately 17-fold higher than CrUBC3 mRNA). Upon heat stress, the two transcripts produce diametrically different responses. In response to 42°C, CrUBC9 transcripts increased approximately 2.7-fold compared with nonstress conditions. CrUBC3, however, shows a drastic decrease in transcript abundance upon heat stress, with transcript levels being reduced to a level approximately 52-fold lower than under nonstress conditions (Supplemental Fig. S3). The opposite response of CrUBC9 and CrUBC3 transcript levels to heat stress strongly suggests that CrUBC9 and CrUBC3 perform distinct functions under stress and nonstress conditions, respectively. In addition, the increase in CrUBC9 transcripts in response to heat stress is consistent with the key role that CrUBC9 plays in this and other stress responses as described below.
mut5 Lacks the Ability to SUMOylate Proteins under a Variety of Stress Conditions
Similar to SUMOylation patterns observed during stress imposed by the 42°C heat treatment, C. reinhardtii also modifies high-Mr proteins with SUMO in response to salt, osmotic, and 37°C stress conditions (Wang et al., 2008). To determine if the SUMOylation deficiency observed in mut5 occurs for all of these abiotic stress treatments, wild-type and mut5 cells were exposed to these and other stress conditions and analyzed by immunoblot analysis with anti-SUMO antibodies. In response to heat stress at 37°C, salinity stress with 175 mm NaCl, osmotic stress with 300 mm sorbitol, and reactive oxygen species stress with 2 mm hydrogen peroxide (H2O2), wild-type cells showed SUMOylation of high-Mr proteins in a pattern similar to that observed with incubation at 42°C (Supplemental Fig. S4). In contrast, under these four additional abiotic stress conditions, mut5 cells showed no change in SUMOylation patterns between nonstressed and stressed conditions, indicating that CrUBC9 is required for stress-induced SUMOylation in response to a variety of environmental adversities.
To determine if CrUBC9 is involved in SUMOylation in response to carbon deprivation, SUMOylation patterns were determined in wild-type and mut5 cells deprived of a carbon source by placing them in the dark and removing acetate from the growth medium. However, attempts to deprive cells of all carbon by placing them in Tris-acetate phosphate (TAP) medium lacking acetate and incubating in the dark proved rapidly lethal for cells. Therefore, to slow the rate of cell death and slow the rate of carbon deprivation, cells were placed in TAP medium containing one-half the normal concentration of acetate and incubated in the dark. Within 48 h, wild-type cells grown in the dark in the presence of medium containing one-half-strength acetate exhibited SUMOylation of several medium to high-Mr proteins (Supplemental Fig. S5). In contrast, mut5 cells incubated in a similar manner displayed no change in SUMOylation pattern. A control culture incubated in full-strength TAP medium in the dark, likewise, showed no change in SUMOylation pattern, indicating that the changes observed in wild-type cells were the result of carbon deprivation and not simply the result of dark incubation (Supplemental Fig. S5, last two lanes). An unexplored point of interest is that the pattern of SUMOylation observed with carbon (acetate) starvation is distinct from those observed under other abiotic stresses. Together, the accumulated results suggest that CrUBC9 likely is responsible for most, if not all, SUMOylation observed in C. reinhardtii in response to stress.
Lack of a Functional CrUBC9 Results in Growth Defects under Stress Conditions
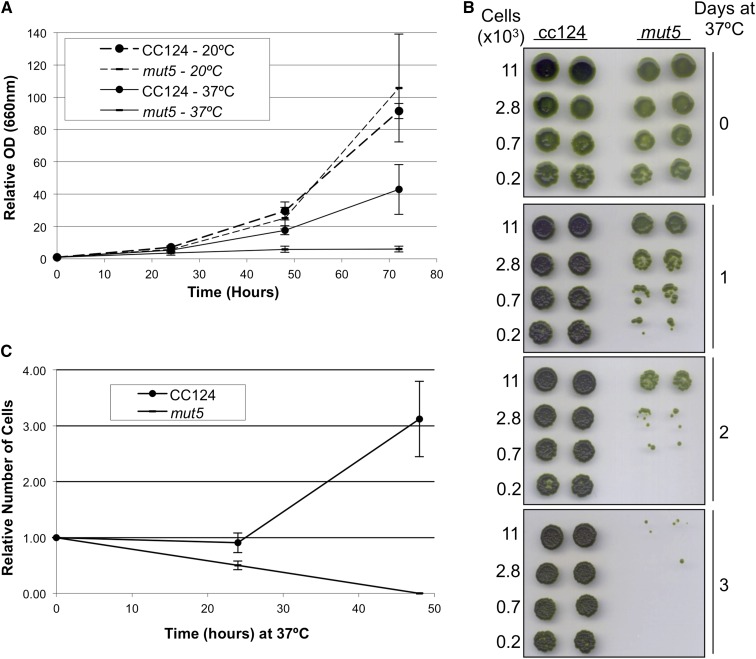
Given that mut5 can grow and SUMOylate the same set of proteins as wild-type cells at 25°C but is incapable of SUMOylation under the stress conditions tested above, it was possible that a growth defect phenotype might be manifested under stress conditions in the absence of CrUBC9. To test this possibility, the growth of wild-type and mut5 cultures in liquid medium was compared at 20°C and 37°C, a temperature known to induce SUMOylation in C. reinhardtii (Supplemental Fig. S4). Dilute cultures of wild-type and mut5 were placed at either 20°C or 37°C, and their growth was monitored over the course of 72 h (Fig. 4A). Growth curves for both wild-type and mut5 cells were similar at 20°C, indicating no deleterious effect on cell viability or growth in mut5 under nonstress conditions. Growth of wild-type cells incubated at 37°C lagged behind both wild-type and mut5 cultures grown at 20°C but still showed increased cell numbers over the course of 72 h. In contrast, mut5 failed to show significant growth at 37°C, only reaching an optical density at 660 nm (OD660) 6 times higher than its initial OD660 by the end of 72 h. The lack of growth observed for mut5 at 37°C raised the possibility that extended exposure to elevated temperatures might prove lethal in cells lacking CrUBC9. To test this conjecture, cultures normalized for cell densities were spotted in a 1:4 dilution series on multiple plates containing TAP medium and incubated at 37°C. Individual plates were removed to room temperature after 24, 48, and 72 h at 37°C (Fig. 4B) and allowed to incubate for an additional 4 to 6 d at room temperature to assess cell viability. The spot tests revealed that wild-type cells maintained on a TAP plate at 37°C for 72 h showed no apparent loss in cell viability (Fig. 4B, rows 1 and 2). However, after just 24 h at 37°C, a loss of viability was apparent with mut5 cells. Loss in viability increased in severity over the course of 72 h to the point that virtually no viable mut5 cells remained after 72 h (Fig. 4B, 3). To better estimate how quickly mut5 cells die at 37°C, very dilute (400 cells per 1 mL) cultures of CC124 and mut5 were again shifted to 37°C, and 250-µL aliquots from these cultures were plated on TAP plates at 24 and 48 h after the shift. The number of colony forming units arising on each plate was taken as a count of the number of living cells in that culture (Fig. 4C). Wild-type cells again showed marked growth over 2 d at 37°C. However, within 24 h at 37°C, the number of colony forming units for mut5 cells had decreased by one-half, and no viable cells were detected at 48 h. The sensitivity of mut5 cells to incubation at 37°C combined with its failure to SUMOylate high-Mr proteins in response to heat stress strongly suggest that the ability of wild-type cells to survive at 37°C is dependent upon SUMOylation of target proteins by CrUBC9.
Figure 4.
mut5 is not viable at 37°C. A, Growth of wild-type and mut5 cultures at 20°C and 37°C was monitored every 24 h for 72 h and reported as OD660 relative to the starting OD660 for each culture. Data points are the average of three independent cultures, and error bars represent the sd. B, A 1:4 dilution series of wild-type and mut5 cells was spotted in duplicate lanes on four individual TAP plates. One was incubated at 25°C, whereas the remaining three were incubated at 37°C in the light. Plates were shifted from 37°C to 25°C at the indicated number of days. C, Wild-type and mut5 cultures were diluted to a concentration of 400 cells mL−1; 250-μL aliquots were plated from initial dilutions and then every 24 h for 48 h after shifting to 37°C. Data points reflect the relative number of cells for each aliquot based on the original dilution and are the average of three independent experiments. Error bars represent the sd.
To determine potential phenotype changes in mut5 when placed under salinity or osmotic stress, the growth of mut5 cells was compared with wild-type cells in the presence of NaCl or sorbitol, respectively. Normalized cultures were spotted in a 1:4 dilution series on TAP plates and TAP plates supplemented with 300 mm sorbitol or 175 mm NaCl. When 1:4 dilution series of wild-type and mut5 cells were spotted on TAP plates, similar growth could be observed between the two, consistent with the growth in liquid culture (Supplemental Fig. S6, A and B, top). However, when those same cells were spotted on TAP + 300 mm sorbitol plates, both wild-type and mut5 cells were capable of growth, but mut5 exhibited a reduced growth rate compared with wild-type cells (Supplemental Fig. S6A). When wild-type and mut5 cells were spotted on medium containing 175 mm NaCl, wild-type cells showed strongly reduced growth compared with control cells spotted on a TAP plate. In contrast, mut5 cells were incapable of growth on plates containing 175 mm NaCl (Supplemental Fig. S6B). Together, these data strongly suggest that SUMOylation of proteins by CrUBC9 enables C. reinhardtii cells to tolerate fluctuations in both their temperature and osmotic environments. Without these protein modifications, survival of C. reinhardtii cells under these conditions becomes impossible or severely compromised.
The CrUbc9 Gene Complements the mut5 Mutant Phenotype
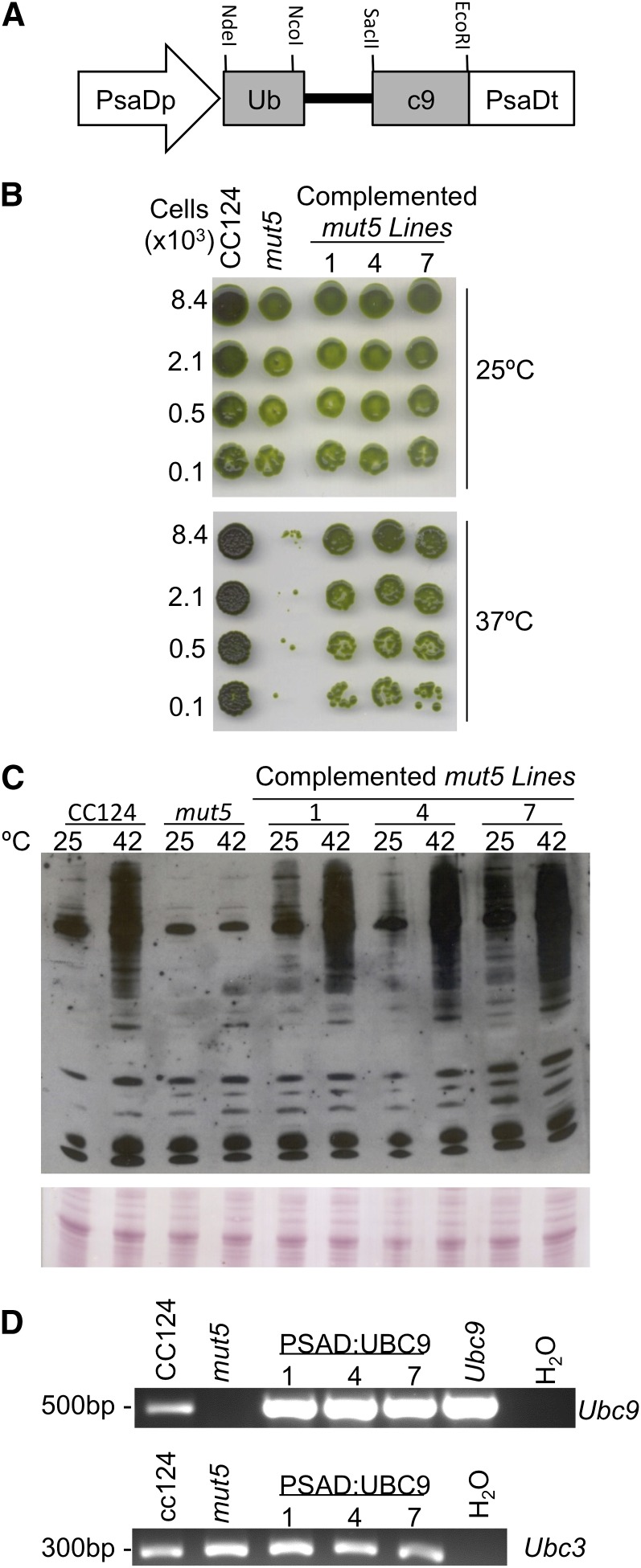
To show that the lack of SUMOylation and other phenotypes observed in mut5 is specifically the result of the CrUBC9 deletion, complementation of mut5 with a CrUBC9 transgene was attempted. The inability of mut5 cells to grow at 37°C was used to screen for complemented cells that were able to grow at this temperature after transformation with a plasmid containing a CrUBC9 transgene (Fig. 5A). mut5 cells were cotransformed with a plasmid containing a CrUBC9 complementary DNA (cDNA) modified to include a single CrUBC9 intron and a plasmid containing the Streptomyces hygroscopicus aminoglycoside phosphotransferase7 gene that confers resistance to hygromycin B (Berthold et al., 2002). Hygromycin B-resistant transformants were spotted on TAP plates and incubated at 37°C to assess their growth. Three lines that appeared to grow at 37°C were rescreened and compared with wild-type and mut5 controls to confirm their phenotypes (Fig. 5B). Not only was growth at 37°C restored, but all three transformants also showed renewed ability to support SUMOylation of high-Mr proteins at 42°C (Fig. 5C). Expression of the newly introduced CrUBC9 gene was confirmed by reverse transcription (RT)-PCR of isolated RNA (Fig. 5D). Complemented lines also showed restored ability to grow on TAP medium supplemented with 300 mm sorbitol and SUMOylate high-Mr proteins in response to carbon deprivation (Supplemental Fig. S7). Combined, these results confirm that the deletion of CrUBC9 in mut5 is responsible for the lack of SUMOylation observed in response to various stress conditions as well as the associated phenotypes.
Figure 5.
Complementation of mut5 with CrUBC9. A, Diagram of the pGenD-Ubc9.int2 expression cassette. Shown are NdeI and EcoRI sites used for cloning CrUbc9 cDNA between the C. reinhardtii PsaD gene promoter and terminator regions in the pGenD vector as well as endogenous NcoI and SacII sites used for adding the second intron of the CrUBC9 gene. B, mut5 lines 1, 4, and 7 putatively complemented with the CrUBC9 gene construct pGenD-Ubc9.int2 were screened for their ability to grow at 37°C. Normalized cell cultures of wild-type, mut5, and complemented lines were spotted on two TAP plates, one of which was incubated at 25°C as a control and one of which was incubated at 37°C for 3 d before shifting to 25°C to assess growth. C, Putative complemented lines were tested for the ability to SUMOylate proteins in response to 42°C. Cell cultures were shifted to 42°C for 1 h, and whole-cell extracts were analyzed for SUMOylation by immunoblot with anti-SUMO antibodies. D, Confirmation of the expression of CrUBC9 in complemented lines by RT-PCR analysis. RNA was isolated from wild-type, mut5, and complemented lines. RT-PCR using CrUbc9-specific primers was used to detect the production of CrUbc9 transcripts. The next to last lane contains the product from RT-PCR amplification of a cDNA clone of CrUBC9. Expression of CrUbc3 was used as a control. Lanes marked H2O are negative controls with no template RNA added.
Localization of CrUBC9
Complementation of mut5 with a chimeric transgene encoding the CrUBC9 protein fused to the mCherry fluorescent protein through a flexible linker region consisting of four repeats of the amino acid sequence Glu-Ala-Ala-Arg (4XEAAR; Elrouby and Coupland, 2010; Supplemental Fig. S8A) was used to determine the in situ localization of CrUBC9 in living cells. The complemented mut5 mutant displayed partially restored growth at 37°C (Supplemental Fig. S8B) and the ability to SUMOylate high-Mr proteins in response to heat shock treatment (Supplemental Fig. S8C). Examination of these cells by confocal microscopy showed a pattern consistent with nuclear localization of CrUCB9 at 25°C as well as during increasing times of exposure to 42°C (Supplemental Fig. S9). Examination at a higher magnification (Fig. 6, A and B) showed that, whereas the preponderance of CrUBC9 resided in the nucleus, trace amounts could be detected in the cytoplasm. This is compared with a control culture expressing mCherry alone, in which the mCherry signal is readily detected in both the nucleus and the cytoplasm (Fig. 6C).
Figure 6.
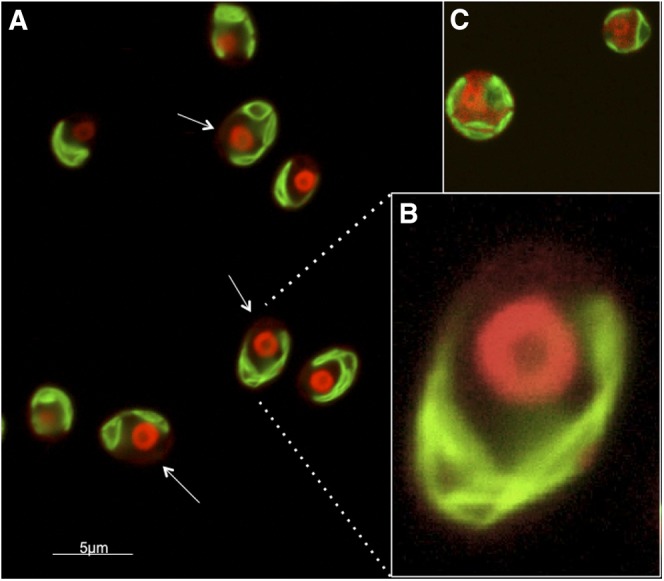
CrUBC9 localizes primarily to the nucleus. A, Fluorescence microscopy image of mut5 cells complemented with CrUBC9-4XEAAAR-mCherry and incubated at 42°C for 60 min. For this image, the mCherry signal was amplified to detect weak signals. White arrows indicate trace mCherry signals that reside outside the nucleus and do not merge with chlorophyll autofluorescence (false green color) from the chloroplast, indicating a cytoplasmic localization. B, Enlarged image of a single cell. C, Image of control cells expressing mCherry lacking the CrUBC9-4XEAAAR tag.
DISCUSSION
Initial characterization of the SUMOylation system in C. reinhardtii included the identification of a putative SUMO E2 conjugase (CrUBC9) that showed in vitro SUMOylation activity (Wang et al., 2008). SUMOylation has been implicated in many important cellular processes, and in a variety of taxonomic groups, SUMO or SUMO pathway mutants are lethal (Seufert et al.,, 1995; Hayashi et al., 2002; Nacerddine et al., 2005; Saracco et al., 2007). There are two reports of viable SUMOylation mutants in S. pombe and Aspergillis nidulans; however, they both exhibit severe growth defects, including reduced cell growth and abnormal cell morphology (al-Khodairy et al., 1995; Wong et al., 2008). The role of SUMOylation in stress response has been shown in both yeast and Arabidopsis in experiments in which the overexpression or heterologous expression of Ubc9 enzymes resulted in increased stress tolerance (Hiraishi et al., 2006; Karan and Subudhi, 2012). Therefore, discovery of mut5, a mutant completely lacking the previously identified SUMO conjugase CrUBC9, was unexpected.
mut5 displayed no obvious growth defect, and under nonstress conditions, mut5 grew at a rate comparable with wild-type cells (Fig. 4A). However, unlike wild-type cells subjected to various stress conditions, mut5 failed to SUMOylate any of a large set of high-Mr proteins when placed in the same stress conditions (Fig. 1; Supplemental Figs. S4 and S5). Moreover, although wild-type cells readily survived the stress conditions, mut5 displayed little or no tolerance to all the stress conditions tested (Figs. 4 and 5; Supplemental Fig. S6). These results combined with our previous demonstration that CrUBC9 has SUMO E2 conjugase activity in vitro (Wang et al., 2008) and the observation that complementation of mut5 with a CrUBC9 transgene restored SUMOylation in response to various stresses (Fig. 5, B and C; Supplemental Fig. S7) provide definitive evidence that CrUBC9 is a functional SUMO E2 conjugase in C. reinhardtii and moreover, uniquely a stress-specific SUMO E2 conjugase, a phenomenon not previously observed in other organisms. The fact that SUMOylation of proteins is essential for survival of C. reinhardtii under harsh conditions bolsters the hypothesis that the extensive stimulation of protein SUMOylation observed in numerous other eukaryotes under stress conditions (Saitoh and Hinchey, 2000; Kurepa et al., 2003; Šramko et al., 2006), likewise, is essential to their survival.
The ability to use a CrUBC9 gene fused with an mCherry coding region for complementation of the growth defect of mut5 at 37°C allowed us to determine the subcellular localization of CrUBC9. Fluorescent confocal microscopy (Fig. 6) showed distinct and nearly exclusive localization of CrUBC9 to the structurally distinct nucleus of C. reinhardtii. This observation complements our earlier studies that used anti-SUMO polyclonal antibodies to detect SUMOylated proteins in the nuclei of fixed cells (Wang et al., 2008). UBC9 homologs identified in other organisms also localize to the nucleus as do proteins targeted for SUMOylation in response to heat stress (Rodriguez et al., 2001; Bruderer et al., 2011).
Our results show that CrUBC9 is categorically required for SUMOylation in response to stress in C. reinhardtii but apparently dispensable under normal growth conditions. This is evidenced by the lack of any observable negative phenotypes in mut5 compared with wild-type cells under nonstress conditions (Fig. 4A) as well as the observation that SUMOylation patterns are identical between wild-type and mut5 cells at 25°C (Fig. 1). These observations are intriguing, because virtually every other haploid or diploid organism studied to date has a single essential SUMO E2 conjugase (Saitoh and Hinchey, 2000; Kurepa et al., 2003; Šramko et al., 2006). Competition assay results verified that the proteins detected under nonstress conditions in both wild-type and mut5 cells are, indeed, SUMOylated (Fig. 2), providing convincing evidence that a second SUMO conjugase must exist in C. reinhardtii. The original description of the SUMO system in C. reinhardtii raised the possibility that the genome could contain as many as four SUMO conjugase enzymes, including CrUBC9, but none of the proposed enzymes was more than 54% identical to CrUBC9 (Wang et al., 2008). A deeper analysis comparing verified yeast E2 conjugase sequences with candidate C. reinhardtii SUMO and ubiquitin E2 conjugases in this study (Fig. 3; Supplemental Table S1) points to CrUBC3 as the putative second SUMO E2 conjugase in C. reinhardtii. Marked amino acid sequence differences between CrUBC9 and this second putative SUMO conjugase are perhaps not surprising given the distinctly different roles that each must play in SUMOylation of proteins in C. reinhardtii, with CrUBC9 responsible for SUMOylation under stress conditions and CrUBC3 likely responsible for housekeeping SUMOylation functions needed for cell growth and division under normal nonstress conditions. Future studies will be aimed at providing verification or denial of CrUBC3 as this second SUMO E2 conjugase.
MATERIALS AND METHODS
Chlamydomonas reinhardtii Strains and Growth Conditions
The wild-type strain CC124 was obtained from the Chlamydomonas Genetics Center at the University of Minnesota. mut5 (Δubc9) is a derivative of CC124. Cultures were grown in TAP medium (Harris, 2009) unless otherwise stated.
Stress Treatments
For heat shock experiments in liquid cultures, 25 mL of midlog phase cells were transferred to a prewarmed 125-mL flask at either 37°C or 42°C as indicated in a prewarmed rotary shaker with continuous light (30–40 μmol s−1 m−2). For high-salt treatment in liquid, TAP medium supplemented with 1.75 m NaCl was added to liquid culture to a final concentration of 175 mm NaCl. For high-osmotic pressure treatment in liquid, TAP medium supplemented with 3 m sorbitol was added to liquid cultures to a final concentration of 300 mm. For H2O2 treatment, H2O2 was added to liquid cultures to a final concentration of 2 mm.
For growth curves at 20°C and 37°C, wild-type and mut5 cultures were diluted in TAP medium to an OD660 between 0.005 and 0.022 and incubated at either 20°C or 37°C under continuous light (30–40 μmol s−1 m−2). OD660 readings were taken every 24 h for 72 h. For growth at 37°C on plates, cultures with normalized cell counts were spotted in a 1:4 dilution series on TAP plates. After incubation at 37°C for the indicated time, plates were shifted to 25°C for several days to assess growth. Growth was compared with a control plate that was maintained at 25°C. For quantification of cell viability at 37°C, midlog phase cells were diluted to a density of 400 cells per 1 mL, so that a 250-μL aliquot of cells would contain approximately 100 cells; 250-μL aliquots of cells from the original dilution as well as after 24 and 48 h at 37°C were plated on TAP plates and incubated at 25°C. The number of colonies arising from each 250-μL aliquot was counted as a measure of the number of viable cells in each 250-μL aliquot. All growth on plates under nonstress (25°C) conditions was in continuous light (100 μmol s−1 m−2).
For cell viability under salt and osmotic stress, normalized cell cultures were spotted in a 1:4 dilution series on TAP and TAP + 175 mm NaCl plates or TAP and TAP + 300 mm sorbitol plates, respectively. Plates were incubated at 25°C (100 μmol s−1 m−2) to assess growth.
For carbon deprivation, cultures were harvested at 2,000g for 5 min and washed two times with sterile water. Final cell pellets were resuspended in equal parts of TAP medium lacking acetate and TAP liquid medium, resulting in a final concentration of 8.7 mm acetate instead of the usual 17.4 mm acetate found in full-strength TAP medium. Cultures were wrapped in aluminum foil to prevent photosynthesis. Control cultures of wild-type cells were resuspended in full-strength TAP medium and also wrapped in aluminum foil to eliminate light. The density of starting cultures was determined by counting cells with a hemacytometer and measuring the OD660. The OD660 of each culture was also determined at 24 and 48 h of nutrient deprivation. The fold increase or decrease in the OD660 compared with that of the initial culture was used to load lysates from equivalent numbers of cells per lane on a BisTris SDS-PAGE gel used for immunoblot analysis.
Immunoblot Analysis
Whole-cell extracts were prepared by lysing cells directly in loading buffer. Cell pellets were resuspended in one-tenth-volume of loading buffer (50 mm Tris-Cl, pH 6.8, 2% [w/v] SDS, 10% [v/v] glycerol, 0.1% [w/v] Bromophenol Blue, and 100 mm β-mercaptoethanol) and boiled for 4 min. Proteins were separated on an 8.5% (w/v) BisTris SDS-PAGE gel in 1× MOPS buffer (50 mm MOPS, 50 mm Tris, 1 mm EDTA, 0.1% [w/v] SDS, and 5 mm sodium bisulfite). After separation, proteins were transferred to a nitrocellulose membrane using a Trans-Blot SD Semi-Dry Transfer Cell (20 V for 1 h; Bio-Rad). After transfer, blots were stained with a solution of 0.05% (w/v) Reactive Brown in 5% (v/v) acetic acid to visualize the proteins transferred to the membrane and assess the loading between lanes. The membrane then was treated with 3% (w/v) milk powder in Tris-buffered saline plus Tween 20 (TBST; 500 mm NaCl, 100 mm Tris-Cl, pH 7.5, and 0.05% [v/v Tween 20) for 30 min at room temperature before incubation in primary antibody overnight at 4°C. After exposure to primary antibody, blots were washed two times in TBST before incubation in secondary horseradish peroxidase (HRP)-tagged donkey anti-rabbit antibody (GE Healthcare) in 1× TBST for 1 h at room temperature. After incubation with secondary antibody, blots were washed two times with TBST and one time with Tris-buffered saline. Protein bands bound by primary antibodies and adherent HRP-labeled antibodies were detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce). The anti-SUMO primary antibody is a polyclonal antibody directed against the CrSUMO96 protein of C. reinhardtii and has been previously described (Wang et al., 2008). A 1:1,000 dilution of the third bleed was used for detection of SUMO proteins. Antibodies to inorganic carbon accumulation mutant5 protein have been previously described (Wang et al., 2005) and were used at a 1:10,000 dilution. Secondary antibody for anti-SUMO and anti-Cia5 primary antibodies were HRP-linked donkey anti-rabbit Igs used at a 1:10,000 dilution.
Competition Assay
Recombinant CrSUMO96 was overexpressed in Escherichia coli and purified as previously described (Wang et al., 2008). For production of CrUBIQ1A, total RNA was isolated from CC124 cells using Trizol LS (Invitrogen) according to the manufacturer’s recommendations. UBIQ1A mRNA was amplified from CC124 total RNA using the following primers: 5′-CCCCCATATGCAGATTTTCGTGAAGACC-3′ (NdeI site underlined) and 5′-CCCAGAATTCAGCCAACGTCCTTCAGC-3′ (EcoRI site underlined). UBIQ1A cDNA was cloned into pET-28b using NdeI and EcoRI restriction sites. UBIQ1A was overexpressed and purified using the same conditions as noted above for CrSUMO96. Proteins were quantified using the Bio-Rad Protein Quantification Kit (Bio-Rad).
For assays testing for the specificity or nonspecificity of SUMO antibody binding to proteins on immunoblots, proteins in equivalent amounts of extracts of CC124 and mut5 from both 25°C and 1 h at 42°C were separated by electrophoresis on three 8.5% (w/v) BisTris SDS-PAGE gels along with 0.5 ng of CrSUMO96 and CrUBIQ1A. Anti-SUMO primary antibodies were preincubated in a dry milk blocking solution alone, blocking solution with 50 μg of recombinant CrSUMO96, or blocking solution with 50 μg of recombinant CrUBIQ1A for 1 h at room temperature before application to one of the blots. After incubation at 4°C overnight in primary antibody solutions, blots were processed as described above for other types of immunoblots.
Cloning of CrUbc9 and Complementation of mut5
The CrUBC9 cDNA (500 bp) was amplified by RT-PCR from C. reinhardtii RNA using the following primers: 5′-TAAACATATGATGTCTGGCGTCGCA-3′ and 5′-AAATGAATTCTCACGAGGGTGGC-3, which added NdeI and EcoRI sites (underlined), respectively. The cDNA was then cloned into the pGenD expression cassette using the NdeI and EcoRI restriction sites (Fischer and Rochaix, 2001). This placed the UBC9 cDNA under the control of the photosystem I gene D (PsaD) promoter. Subsequently, the second intron plus some of the flanking coding sequence of the CrUBC9 gene was amplified from C. reinhardtii genomic DNA using the following primers: 5′-GAACCTGATGAAGTGGAAGTGCC-3′ and 5′-TAGATGTTGGGGTGGAAGAAGC-3′. The resulting genomic DNA fragment contained an endogenous NcoI site in the coding region flanking the intron at the 5′ end and an endogenous SacII site in the coding region flanking the intron at the 3′ end. These restriction sites were used to clone the intron into the CrUBC9 cDNA. The resulting plasmid was named pGenD-Ubc9.int2.
For complementation, mut5 cells were cotransformed with pPsaD-Ubc9.int2 and pHyg3, which confers resistance to Hygromycin B (Berthold et al., 2002), using standard electroporation conditions (Shimogawara et al., 1998). Transformants were selected on medium containing 30 μg mL−1 Hygromycin B. Individual transformants were picked into 100 μL of TAP medium in the first and fourth rows of a 96-well plate and grown for 24 to 48 h at 25°C. The second, third, and fifth through eighth rows of the same plate were used to generate a 1:4 dilution series of each transformant that was subsequently spotted on two individual TAP plates. One plate was incubated at 25°C, whereas the other was incubated at 37°C for 3 d. Colonies that showed growth at 37°C after 3 d were rescreened in a similar dilution series assay to assess growth of putative complemented cells at 37°C compared with wild-type and mut5 cells. Expression of CrUbc9 mRNA was confirmed in complemented lines by RT-PCR using the SuperScript III One-Step RT-PCR System (Invitrogen) and the same primers described above that were used for the cloning of the CrUBC9 cDNA. As a control for the presence of mut5 RNA, a portion of a second predicted UBC transcript, CrUbc3, was amplified using the following primers: 5′-TACGGCTTTGTGGGGAAACCATTGACC-3′ and 5′-ACGATCTGCACGATGTTGATGCTGG-3′.
CrUBC9-4XEAAAR-mCherry Fusion
For expression of a CrUBC9-4XEAAAR-mCherry fusion, CrUBC9 cDNA from the pGenD-Ubc9.int2 plasmid was amplified with the following primers: 5′-GAACCTGATGAAGTGGAAGTGCC-3′ and 5′-TAAAAGATCTCGAGGGTGGCGGG-3′ (BglII site underlined). A segment of DNA encoding four repeats of Glu, Ala, Ala, Ala, and Arg (4XEAAAR) was generated by annealing the following primers and filling in with Phusion DNA Polymerase (Thermofisher Scientific): 5′-TCAAGATCTGAGGCCGCTGCCCGCGAGGCTGCCGCTCGGGAGGCGGCT-GCCCGC-3′ and 5′-CTGCAGGTCGACTCTAGCGCGGGCCGCAGCCTCGCG-GGCAGCCGCCTC-3′. The resulting double-stranded fragment encoded four EAAAR repeats flanked by in-frame BglII and SalI restriction sites. mCherry cDNA was amplified using the following primers: 5′-TAAAGTCGACGTGAGCAAGGGCGAGG-3′ (SalI site underlined) and 5′-GATGAATTCTTAGTAGGTACCGTTGCTAGCCTTGTACAGCTCGTCCATG-3′ (EcoRI site underlined.) The corresponding BglI sites in the amplified CrUBC9 cDNA and 4XEAAAR fragment and SalI sites in the 4XEAAAR fragment and mCherry cDNA were used to ligate the three fragments together to generate the CrUBC9-4XEAAAR-mCherry fusion. The previously described endogenous NcoI site of the CrUBC9 cDNA and EcoRI site of the mCherry cDNA were used to ligate the fusion into pGenD-Ubc9.int2. The resulting plasmid was named pUxM.
Confocal Microscopy
Live images of C. reinhardtii were captured using a Nikon A1 confocal imaging system mounted on a Nikon Eclipse 90i microscope with a 100× objective. mCherry fluorescent signal was acquired with 561.5-nm excitation and 570- to 620-nm emission, and chlorophyll autofluorescence signal was acquired at 641-nm excitation and 662- to 737-nm emission and pseudocolored green for visualization. Control cells expressing free mCherry were generated as previously described (Rasala et al., 2013).
Bioinformatic Analysis of SUMO Conjugase Candidates in C. reinhardtii
To identify potential additional SUMO conjugase candidates, an alignment was generated with CrUBC9 (Cre02.g142000), CrUBC3 (Cre03.g167000), Cre16.g693700, and Cre06.g292800 using sequences deposited in Phytozome (http://phytozome.jgi.doe.gov/) and known SUMO conjugase and ubiquitin conjugase enzymes from yeast (Saccharomyces cerevisiae). Yeast enzymes included the SUMO conjugase ScUBC9 (P50623.1) and the ubiquitin conjugase enzymes ScUBC2 (P06104.1), ScUBC4 (P15731.15), ScUBC5 (P15732.1), ScUBC7 (Q02159.1), ScUBC11 (P52492.1), and ScUBC12 (P52490.1). The only ubiquitin conjugase enzymes excluded were those with a length drastically different from the C. reinhardtii candidate proteins. The alignment was generated using ClustalOmega (Sievers et al., 2011).
To generate a phylogenetic tree, an alignment of the top homologs in the UniProt/SwissProt database for each of four putative SUMO conjugases from C. reinhardtii as well as selected other E2 conjugase sequences, including ubiquitin fold modifier1 E2 conjugases, Related to ubiquitin1/neural precursor cell expressed, developmentally down8 E2 conjugases, SUMO E2 conjugases, and Ubiquitin E2 conjugases, was generated using MUSCLE (Edgar, 2004). Gaps in the alignment were removed using GBLOCKS (Castresana, 2000). A tree was generated from the resulting alignment with PhyML (Guindon et al., 2010) using the substitution model LG (Le and Gascuel, 2008) and performing bootstrap analysis on 100 replicates. Annotation of the tree and highlighting of selected clusters were performed using MEGA 6 (Tamura et al., 2013).
Quantitation of Expression Levels of CrUBC9 and CrUBC3
For quantitation of expression levels of CrUbc9 and CrUbc3 transcripts, total RNA was isolated from CC124 C. reinhardtii cells grown at 25°C and after a shift to 42°C for 1 h using Trizol LS (Invitrogen) according to the manufacturer’s recommendations. Contaminating DNA was removed by treatment with DNaseI (ThermoScientific). cDNA was synthesized from 2.4 μg of total RNA with oligo(dT) primers using the Plexor Two-Step qRT-PCR System (Promega). Synthesized cDNA was diluted 1:2 in 1 mm MOPS:1 mm EDTA and used as substrate for a quantitative PCR reaction according the Plexor Two-Step qRT-PCR System recommendations using 0.2 mm each primer (Biosearch Technologies). Primer pairs for quantitative PCR were as follows: CrUBC3, 5′-FAM-isoC-GCTGGGGTACACGTTTGGATG-3′ and 5′-GATACCAGGGCCGGAGAAGAC-3′; CrUBC9, 5′-CAL Fluor Orange 560-isoC-TGAGGCACACGGTACCGGAG-3′ and 5′-CTCACCATGGAGTTCAGCGAG-3′; and G-Protein, 5′-Quasar 670-isoC-GTTGGTGGTCATGGGCGAGAA-3′ and 5′-GACAAGACCATCAAGCTGTGGAAC-3′. G-Protein and CrUBC9 primers were multiplexed in a single reaction. The efficiency of amplification for each set of primers was calculated and used to quantify the relative transcript abundance relative to CrUBC9 at 25°C (Pfaffl, 2001). Efficiencies for the primer pairs were as follows: CrUBC9, 83%; CrUBC3, 82%; and G-Protein, 82%.
Sequence data for this article are listed in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. mut5 is unable to SUMOylate stress-related proteins during prolonged heat stress at 42°C.
Supplemental Figure S2. Amino acid sequence alignments comparing verified yeast SUMO and ubiquitin E2 conjugases with putative SUMO and ubiquitin E2 conjugases from C. reinhardtii.
Supplemental Figure S3. Comparison of CrUBC9 and CrUBC3 mRNA levels in wild-type cells in response to heat shock.
Supplemental Figure S4. mut5 fails to SUMOylate high-Mr proteins in response to diverse abiotic stresses.
Supplemental Figure S5. mut5 fails to SUMOylate high-Mr proteins in response to carbon source deprivation.
Supplemental Figure S6. Phenotypes of mut5 under osmotic and salt stresses.
Supplemental Figure S7. Phenotypes of complemented mut5.
Supplemental Figure S8. Complementation of mut5 with a CrUBC9-4XEAAAR-mCherry fusion.
Supplemental Figure S9. CrUBC9 localizes to the nucleus at 25°C and 42°C.
Supplemental Table S1. Identification of the closet orthologs to SUMO E2 conjugases and ubiquitin E2 conjugases in yeast and C. reinhardtii.
Supplemental Table S2. Annotation of protein sequences used for generation of the phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Dr. Jeff Mower for generous assistance with phylogenetic analyses of the E2 conjugase families.
Glossary
- cDNA
complementary DNA
- HRP
horseradish peroxidase
- H2O2
hydrogen peroxide
- RT
reverse transcription
- TAP
Tris-acetate phosphate
- TBST
Tris-buffered saline Tween 20
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–1052281 to H.C. and MCB–0952533 to D.P.W.) and the EPSCoR (grant no. EPSCoR–1004094 to H.C. and D.P.W.).
Articles can be viewed without a subscription.
References
- al-Khodairy F, Enoch T, Hagan IM, Carr AM (1995) The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci 108: 475–486 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F (2001) Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W (2002) An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT (2011) Purification and identification of endogenous polySUMO conjugates. EMBO Rep 12: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Chen YY, Yeh KC (2011) Effect of Cu content on the activity of Cu/ZnSOD1 in the Arabidopsis SUMO E3 ligase siz1 mutant. Plant Signal Behav 6: 1428–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A (2010) Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol Biol 74: 33–45 [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JMP, Thomson J, Hay RT (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett 417: 297–300 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Coupland G (2010) Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107: 17415–17420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Rochaix JDR (2001) The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics 265: 888–894 [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT (2009) System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD (2001) Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem 276: 18513–18518 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Harris EH. (2009) The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, Ed 2 Academic Press, Oxford [Google Scholar]
- Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T (2002) Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res 280: 212–221 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraishi H, Mochizuki M, Takagi H (2006) Enhancement of stress tolerance in Saccharomyces cerevisiae by overexpression of ubiquitin ligase Rsp5 and ubiquitin-conjugating enzymes. Biosci Biotechnol Biochem 70: 2762–2765 [DOI] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276: 40263–40267 [DOI] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, et al. (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60: 666–678 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H (2006) Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314: 1893–1898 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H (2010) Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA 107: 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802 [DOI] [PubMed] [Google Scholar]
- Karan R, Subudhi PK (2012) A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biol 12: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107: 16512–16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Vierstra RD (2011) Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signal Behav 6: 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Sato A, Ohta M, Furukawa J (2011) Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta 234: 1191–1199 [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9: 769–779 [DOI] [PubMed] [Google Scholar]
- Nigam N, Singh A, Sahi C, Chandramouli A, Grover A (2008) SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Genet Genomics 279: 371–383 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Tomanov K, Hofmann K, Stuible HP, Bachmair A (2012) Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytol 195: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M, Hammerschmidt M (2006) Ubc9 regulates mitosis and cell survival during zebrafish development. Mol Biol Cell 17: 5324–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Yun DJ (2013) New insights into the role of the small ubiquitin-like modifier (SUMO) in plants. Int Rev Cell Mol Biol 300: 161–209 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Barrera DJ, Ng J, Plucinak TM, Rosenberg JN, Weeks DP, Oyler GA, Peterson TC, Haerizadeh F, Mayfield SP (2013) Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J 74: 545–556 [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Wendel JF (2014) Doubling down on genomes: polyploidy and crop plants. Am J Bot 101: 1711–1725 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654–12659 [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275: 6252–6258 [DOI] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S (1995) Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373: 78–81 [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šramko M, Markus J, Kabát J, Wolff L, Bies J (2006) Stress-induced inactivation of the c-Myb transcription factor through conjugation of SUMO-2/3 proteins. J Biol Chem 281: 40065–40075 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Hateboer G, Perrakis A, Bernards R, Sixma TK (1997) Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J Biol Chem 272: 21381–21387 [DOI] [PubMed] [Google Scholar]
- Villajuana-Bonequi M, Elrouby N, Nordström K, Griebel T, Bachmair A, Coupland G (2014) Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J 79: 206–219 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ladunga I, Miller AR, Horken KM, Plucinak T, Weeks DP, Bailey CP (2008) The small ubiquitin-like modifier (SUMO) and SUMO-conjugating system of Chlamydomonas reinhardtii. Genetics 179: 177–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun Z, Horken KM, Im CS, Xiang Y, Grossman AR, Weeks DP (2005) Analyses of CIA5, the master regulator of the carbon-concentrating mechanism in Chlamydomonas reinhardtii, and its control of gene expression. Can J Bot 83: 765–779 [Google Scholar]
- Wong KH, Todd RB, Oakley BR, Oakley CE, Hynes MJ, Davis MA (2008) Sumoylation in Aspergillus nidulans: sumO inactivation, overexpression and live-cell imaging. Fungal Genet Biol 45: 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Yang C (2013) Emerging role of SUMOylation in plant development. Plant Signal Behav 8: e24727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Howley PM (1996) Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res 24: 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142: 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.