Abstract
Gene expression profiling (GEP), which can divide DLBCL into three groups, is impractical to perform routinely. Although algorithms based on immunohistochemistry (IHC) have been proposed as a surrogate for GEP analysis, the power of them has diminished since rituximab added to the chemotherapy. We assessed the prognostic value of four conventional algorithms and the genes in each and out of algorithm by IHC and fluorescence in situ hybridization in DLBCL patients receiving immunochemotherapy. The results showed that neither single protein within algorithms nor the IHC algorithms themselves had strong prognostic power. Using MYC aberrations (MA) either on the genetic or protein levels, we established a new algorithm called MA that could divide patients into distinct prognostic groups. Patients of MA had much shorter overall survival (OS) and progression-free survival (PFS) than non-MA (2-year OS: 56.9% vs. 98.7%; 2-year PFS: 26.8% vs. 86.9%; P < 0.0001 for both). In conclusions, using additional prognostic markers not associated with cell of origin may accurately predict outcomes of DLBCL. Studies with larger samples should be performed to confirm our algorithm and optimize the prognostic system of DLBCL.
Keywords: Diffuse large B-cell lymphoma, algorithms, immunohistochemistry, fluorescence in situ hybridization, MYC aberrations
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults, accounting for 30%-40% of all cases of non-Hodgkin lymphoma (NHL) in Western countries [1]. The standard therapy for patients with DLBCL is rituximab combined with cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), which has a 10-year disease-free survival (DFS) of approximately 42.6% [2]. Gene expression profile (GEP) studies have confirmed that DLBCL can be subdivided into subtypes depending on their gene signatures: germinal center B-cell (GCB) or GCB-like, activated B-cell (ABC) or ABC-like, and type 3 [3,4]. The molecular and genetic distinction, such as MYC rearrangement concurrent with BCL2 rearrangement [5,6], is also important, for patients in subgroups defined by these features respond differently and have different prognoses when treated with R-CHOP [7].
Because GEP is expensive and not readily available in routine practice, several algorithms have been proposed in recent years; these algorithms have been based on immunohistochemical (IHC) staining or tissue microarray analysis, which is a surrogate for GEP analysis [8]. GCB subtype tend to have a more favorable outcome than those with non-GCB subtype in patients treated with CHOP, irrespective of the International Prognostic Index (IPI) score [9]. However, since rituximab was added to CHOP as the standard of care, different factors have been shown to be important in determining a patient’s prognosis when using different IHC algorithms for prediction [7,10,11].
The most commonly used algorithm, proposed by Hans et al [12] is based on the expression of three proteins (CD10, Bcl6 and MUM1/IRF4) which can classify DLBCL patients into two categories (GCB and non-GCB) with different prognoses. This algorithm, however, was created for use in patients to be treated only with CHOP. In addition, the predictions made by this algorithm had low concordance with those from GEP analysis (71% concordance for GCB, and 88% for non-GCB) [8]. The prognostic relevance of the Hans algorithm led to inconsistent results in subsequent studies performed in patient groups treated with R-CHOP [7,13-18].
In 2009, Choi et al [15] reported a combination of five markers: GCET1, CD10, Bcl6, MUM1/IRF4, and FOXP1, which can achieve a concordance of about 90% with the GEP in patients treated with R-CHOP [15]. Compared with the Hans algorithm, the Choi algorithm integrated the analysis of two new molecules: FOXP1 and GCET1. Prediction was more accurate than that of the Hans algorithm and facilitated risk stratification of DLBCL patients.
In 2011, Meyer et al [19] reported another algorithm (called the “Tally” algorithm) that had a high concordance (93%) with GEP and was also based on the expression of five markers: CD10, GCET1, FOXP1, MUM1, and LMO2. This method includes an equal number of GCB (GCET1 and CD10) and non-GCB (FOXP1 and MUM1) antibodies. Classification is determined by the immunophenotype pair with more positive antigens. If an equal number of GCB and ABC antigens are positive, then LMO2 determines the immunophenotype (i.e., LMO2 ≥ 30% yields GCB).
Recently, a report from the International DLBCL R-CHOP Consortium introduced a new algorithm called “Visco-Young” ; it is based on the expression of CD10, FOXP1, and Bcl6, and it demonstrated a concordance of 92.6% with GEP and the ability to independently predict the rate of progression-free survival (PFS) and overall survival (OS) [8]. In multivariate analysis, both the IPI and the Consortium’s algorithm were significant independent predictors of PFS and OS [8].
It is notable that most of these algorithms (Figure 1) have been developed and tested only in Western countries; few reports have confirmed the practicability of their use in Eastern or other nations. In this study, we analyzed the four algorithms to determine each one’s power to predict the prognosis of Eastern DLBCL patients treated with R-CHOP-like therapies.
Figure 1.
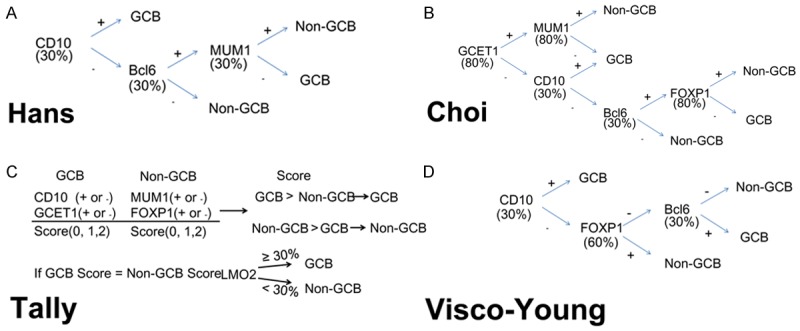
The four algorithms applied in this study were Hans (A), Choi (B), Tally (C), and Visco-Young (D). Abbreviation: GCB: germinal center B-cells.
Material and methods
Ethics statement
All patients provided informed consent in accordance with requirements of the Declaration of Helsinki, and the research project was approved by the University and Institutional Review Boards.
Patients
We retrospectively studied 244 adult patients with de novo DLBCL who had been diagnosed between February 2006 and January 2014. All of the paraffin-embedded sections were reviewed by two hematopathologists (QXG and TXL), and the diagnoses were based on the World Health Organization classification criteria [20]. Among these 244 patients, 141 cases were treated with R-CHOP-like therapy, which was used for prognostic analysis.
IHC
IHC was performed on 4 μm formalin-fixed paraffin-embedded (FFPE) sections. The antibodies used were CD10, Bcl6, MUM1, FOXP1, GCET1, LMO2, Myc, and Bcl2. The cutoff scores for each antibody were described previously [8,12,15,19].
Fluorescence in situ hybridization (FISH)
FISH analysis was performed using FFPE tissue sections according to the manufacturer’s instructions with MYC dual-color, break-apart translocation probe (Vysis LSI) and IGH/BCL2 dual-color, dual-fusion translocation probe (Vysis LSI). The cut-off levels for the probes were established by evaluating the split signal distribution in samples of reactive lymphoid tissues, calculating the mean number of split signals plus three times the standard deviation. The cut-off levels were 14% and 5% for MYC break apart probe and IGH/BCL2 dual-color, dual-fusion translocation probe.
Statistical analysis
The OS and PFS distributions for each algorithm were estimated by the Kaplan-Meier method, with differences evaluated by the log-rank test. OS was defined as the time from initial diagnosis to death or last follow-up. PFS was defined as the time from initial diagnosis to disease progression, start of salvage treatment, additional (unplanned) treatments, relapse, or death from any cause, additional therapy, day of relapse, or day of death from any cause. Patients who were alive and progression-free at last follow-up were censored for this analysis. Chi-squared and Fisher exact tests were used to determine the level of consistency among algorithms and pairwise agreement between different proteins. The Spearman test was used to analyze correlations among variables. For all tests, a P value of 0.05 was considered statistically significant.
Results
Algorithms applied in this study
The published algorithms examined in this study were those of Hans [12], Choi [15], Tally [19], and Visco-Young [8].
According to the algorithms applied in this study, 244 cases of de novo DLBCL could be further investigated by IHC. For the Hans algorithm, 99 cases were classified as GCB and 145 cases as non-GCB. For the Choi, Tally, and Visco-Young algorithms, 119 and 125, 69 and 175, and 107 and 137 cases were classified as GCB and non-GCB types, respectively. Details of the results are shown in Table 1 and Figure 2.
Table 1.
Consistent and inconsistent numbers (percentages) of cases between pairs of algorithms
| Algorithms | H-G | H-non-G | C-G | C-non-G | T-G | T-non-G | V-G | V-non-G |
|---|---|---|---|---|---|---|---|---|
| H-G | / | / | 89 (36.5) | 10 (4.1) | 56 (23.0) | 43 (17.6) | 83 (34.0) | 16 (6.6) |
| H-non-G | / | / | 30 (12.3) | 115 (47.1) | 13 (5.3) | 132 (54.1) | 24 (9.8) | 121 (49.6) |
| C-G | 89 (36.5) | 30 (12.3) | / | / | 57 (23.4) | 62 (25.4) | 107 (43.9) | 12 (4.9) |
| C-non-G | 10 (4.1) | 115 (47.1) | / | / | 12 (4.9) | 113 (46.3) | 0 (0.0) | 125 (51.2) |
| T-G | 56 (23.0) | 13 (5.3) | 57 (23.4) | 12 (4.9) | / | / | 53 (21.7) | 16 (6.6) |
| T-non-G | 43 (17.6) | 132 (54.1) | 62 (25.4) | 113 (46.3) | / | / | 54 (22.1) | 121 (49.6) |
| V-G | 83 (34.0) | 24 (9.8) | 107 (43.9) | 0 (0.0) | 53 (21.7) | 54 (22.1) | / | / |
| V-non-G | 16 (6.6) | 121 (49.6) | 12 (4.9) | 125 (51.2) | 16 (6.6) | 121 (49.6) | / | / |
Abbreviations: GCB: germinal center B-cells; H-G: Hans GCB subtype; H-non-G: Hans non-GCB subtype; C-G: Choi GCB subtype; C-non-G: Choi non-GCB subtype; T-G: Tally GCB subtype; T-non-G: Tally non-GCB subtype; V-G: Visco Young GCB subtype; V-non-G: Visco-Young non-GCB subtype
Figure 2.
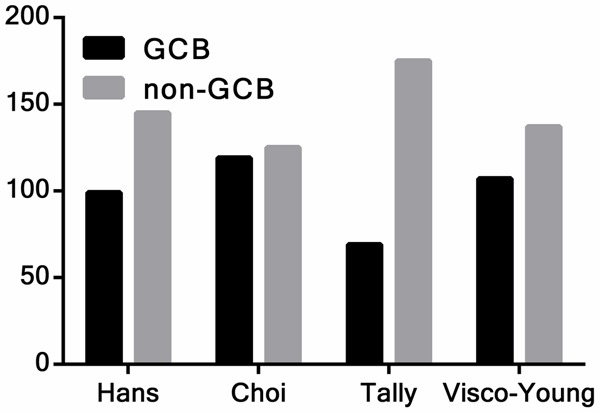
The distribution of GCB and non-GCB patients for each algorithm. There were significant differences among the algorithms (P < 0.0001). Abbreviation: GCB: germinal center B-cells.
The consistency across all four algorithms was 63.52% (155/244). When the results of the algorithms were compared pairwise, however, the consistency was generally better. The Choi and Visco-Young algorithms showed the highest concordance rate (95.08%, x2 = 200.178, κ = 0.901), while the Choi and Tally algorithms had the lowest concordance rate (69.67%, x2 = 44.090, κ = 0.387). The details of the agreements among the algorithms were illustrated in Table 2.
Table 2.
Concordance rates (x2 values) and κ coefficients for the four IHC algorithms
| Algorithm | Hans | Choi | Tally | Visco-Young | |||
|---|---|---|---|---|---|---|---|
| Hans | x2 = 112.789 | κ = 0.671 | x2 = 65.724 | κ = 0.500 | x2 = 108.180 | κ = 0.664 | |
| Choi | x2 = 44.090 | κ = 0.387 | x2 = 200.178 | κ = 0.901 | |||
| Tally | x2 = 42.445 | κ = 0.394 | |||||
P < 0.001 for all x2 analyses. Abbreviation: IHC: immunohistochemistry.
Prognostic significance of original IHC algorithms
The baseline characteristics of the 141 patients in the R-CHOP-like group are listed in Table 3. None of the four algorithms showed significant differences in OS and PFS (except for Tally algorithm, P = 0.022 for PFS) between patients with GCB and non-GCB subtypes (Table 4, Figure 3).
Table 3.
Baseline characteristics of the 141 patients in the R-CHOP-like group
| Characteristic | No. (%) of patients |
|---|---|
| Age | |
| ≤ 60 | 82 (58.2) |
| Male | 92 (65.2) |
| Stage III-IV | 75 (53.2) |
| Elevated LDH | 59 (41.8) |
| PS ≥ 2 | 29 (20.6) |
| Extranodal sites ≥ 2 | 34 (24.1) |
| IPI score of 3-5 | 50 (35.4) |
| B symptoms | 52 (36.9) |
| Treatment responses | |
| CR | 82 (58.2) |
| PR | 35 (24.8) |
| SD/PD | 24 (17.0) |
Abbreviations: PS: Eastern Cooperative Oncology Group performance status; IPI: International Prognostic Index; LDH: lactate dehydrogenase; CR: complete remission; PR: partial remission; SD/PD: stable disease/progression of disease.
Table 4.
Differences in survival for the four tested algorithms between GCB and non-GCB patient subgroups
| Algorithm | Subtype | Numbers | P value (OS) | P value (PFS) |
|---|---|---|---|---|
| Hans | GCB | 62 | 0.142 | 0.210 |
| Non-GCB | 79 | |||
| Choi | GCB | 71 | 0.705 | 0.808 |
| Non-GCB | 70 | |||
| Tally | GCB | 44 | 0.188 | 0.022 |
| Non-GCB | 97 | |||
| Visco-Young | GCB | 64 | 0.706 | 0.889 |
| Non-GCB | 77 |
Abbreviations: GCB: germinal center B-cells; OS: overall survival; PFS: progression-free survival
Figure 3.
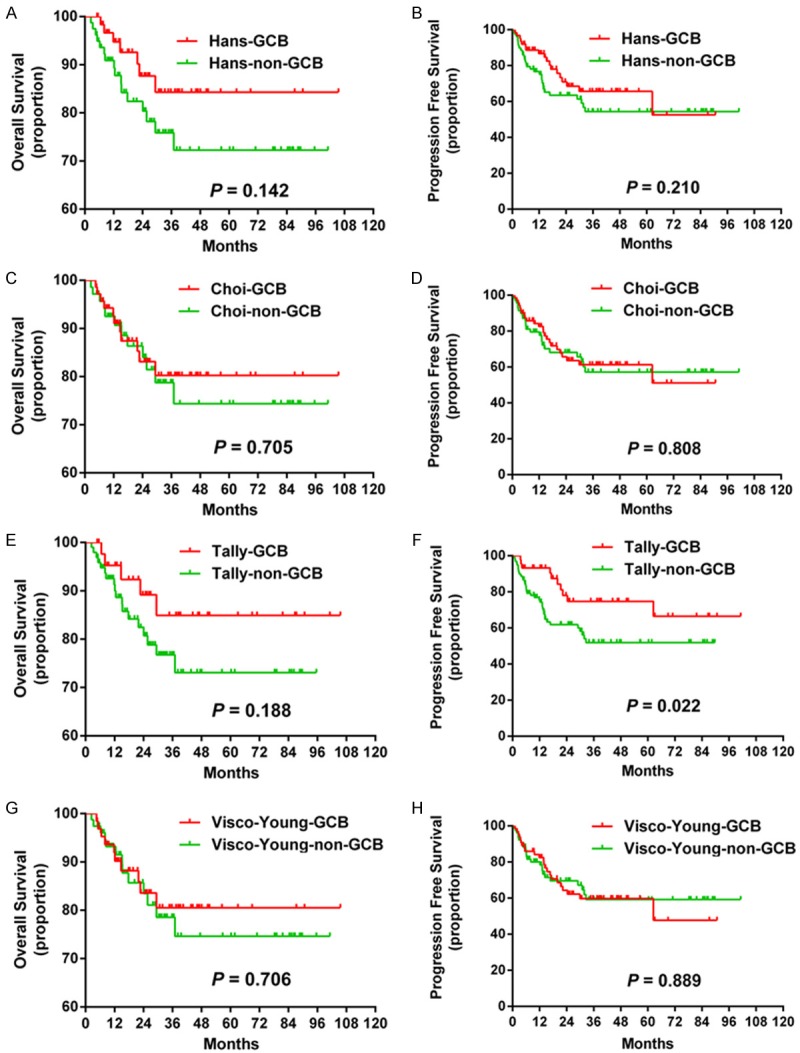
Survival curves calculated using the Hans, Choi, Tally, and Visco-Young algorithms. The Hans Choi and Visco-Young algorithms showed no differences in OS (A, C and G) or PFS (B, D and H). The Tally algorithm showed significant differences in PFS (F) but not OS (E). Abbreviations: GCB: germinal center B-cells; OS: overall survival; PFS: progression-free survival.
Prognostic significance of single markers
Since the four algorithms showed poor prognostic significance, we analyzed single protein in each algorithm (Table 5). None of the proteins predicted significant differences in survival. In addition, pairwise agreement and correlation tests showed that LMO2 had a negative correlation with other GCB markers (data not show).
Table 5.
Prognosis predicted by single protein expression or gene rearrangement
| Variables | No. of patients | P value for OS | P value for PFS |
|---|---|---|---|
| CD10-30% | 37 vs. 94 | P = 0.456 | P = 0.333 |
| Bcl6-30% | 98 vs. 43 | P = 0.621 | P = 0.263 |
| MUM1-30% | 86 vs. 55 | P = 0.183 | P = 0.315 |
| GCET1-30% | 41 vs. 100 | P = 0.632 | P = 0.175 |
| GCET1-80% | 17 vs. 124 | P = 0.387 | P = 0.885 |
| FOXP1-30% | 106 vs. 35 | P = 0.282 | P = 0.128 |
| FOXP1-60% | 84 vs. 57 | P = 0.898 | P = 0.559 |
| FOXP1-80% | 69 vs. 72 | P = 0.710 | P = 0.947 |
| LMO2-30% | 108 vs. 33 | P = 0.587 | P = 0.385 |
| Myc-40% | 42 vs. 99 | P< 0.0001 | P< 0.0001 |
| Bcl2-50% | 72 vs. 69 | P = 0.154 | P = 0.009 |
| MYC rearrangement | 15 vs. 126 | P< 0.0001 | P< 0.0001 |
| BCL2 rearrangement | 20 vs. 121 | P = 0.392 | P = 0.298 |
Abbreviations: OS: overall survival; PFS: progression-free survival.
Furthermore, we observed a cohort of patients, treated with chemoimmunotherpy, whose disease had progressed and who had died mostly in the first two years. In order to determine whether these patients had special poor prognosis factors, we performed IHC and FISH with additional markers. MYC and BCL2, two factors receiving considerable attention currently, were analyzed in our cohort of patients. On the protein level, Myc expression showed significantly decreased survival (2-year OS, 53.4% vs. 96.6%, P < 0.0001; 2-year PFS, 27.5% vs. 81.9%, P < 0.0001). Bcl2 protein, however, predicted significant differences in PFS (2-year PFS, 57.2% vs. 76.9%, P = 0.009) but not OS (2-year OS, 79.4% vs. 90.8%, P = 0.154). On the gene level, the results showed that MYC rearrangement predicted decreased OS (2-year OS, 52.5% vs. 89.1%, P < 0.0001) and PFS (2-year PFS, 33.3% vs. 71.6%, P < 0.0001) (Table 5). However, BCL2 rearrangement showed no differences in survival for either OS ((2-year OS, 74.5% vs. 86.3%, P = 0.392) or PFS (2-year PFS, 48.5% vs. 69.4%, P = 0.298) (Table 5).
Prognostic significance of adding MYC aberrations to the original IHC algorithms
Since the four tested algorithms showed poor predictive ability and the single marker results showed only Myc protein and MYC rearrangement predicted significant outcomes for DLBCL patients. We decided to combine Myc protein and MYC rearrangement into an additional reference index, called “MYC aberrations” (MA). MA defined a single unfavorable group, with either Myc expression or MYC rearrangement. Cases without MA were reclassified using the original algorithms. Each new algorithm was designated by adding “-MA” to the original name (for example, the new Hans algorithm was called Hans-MA in order to distinguish it from the original).
All the four new algorithms showed significant differences in OS and PFS between MA and GCB or non-GCB (Figure 4). However, no differences of OS and PFS (except for the Tally-MA algorithm) were observed between the GCB and non-GCB groups.
Figure 4.
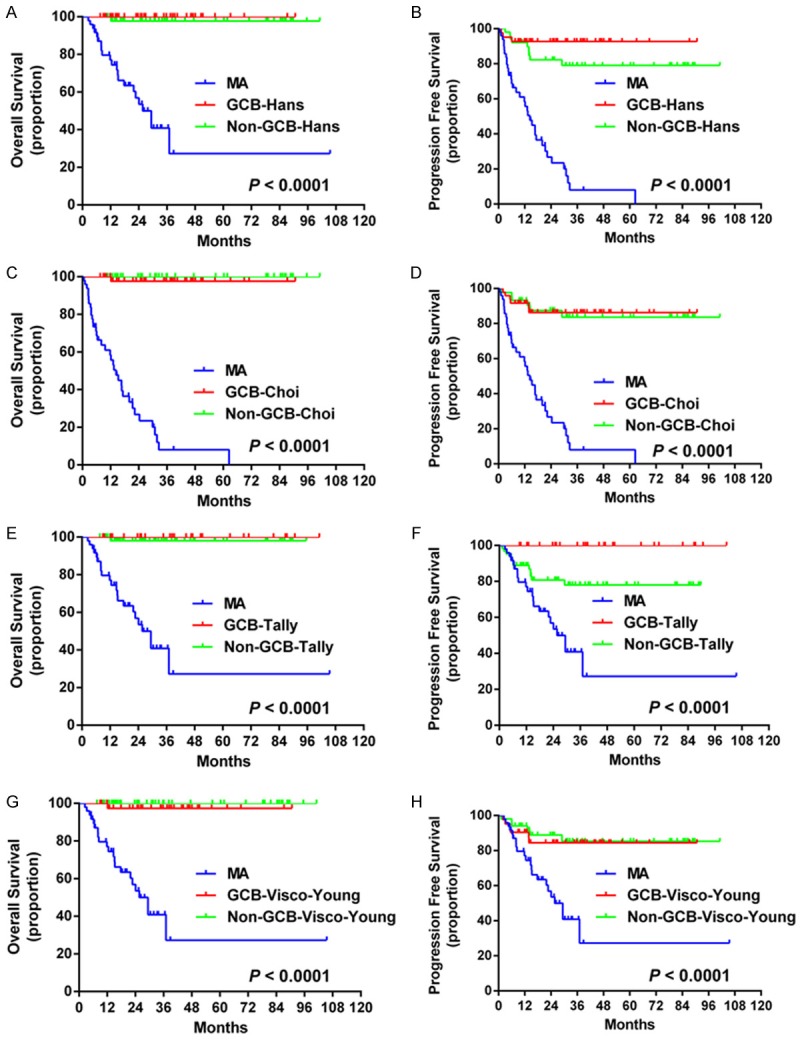
Survival curves for the Hans-MA, Choi-MA, Tally-MA, and Visco-Young-MA algorithms. The algorithms of Hans-MA, Choi-MA and Visco-Young-MA showed significant differences in OS (A, C, G) and PFS (B, D, H) between MA and GCB or non-GCB groups while no differences in OS and PFS were observed between the GCB and non-GCB groups. The Tally-MA algorithm showed significant differences in OS (E) between the MA and GCB or non-GCB groups and in PFS (F) among the three groups (GCB, non-GCB and MA). Abbreviations: GCB: germinal center B-cells; OS: overall survival; PFS: progression-free survival; MA: MYC aberrations.
Prognostic significance of MA
We then used MA as a single algorithm, without including the original algorithms. We defined three MA types. Type 1 was negative for both Myc protein and MYC rearrangement. Type 2 was as positive for either Myc protein or MYC rearrangement. Type 3 was positive for both Myc protein and MYC rearrangement. Survival analysis showed significant differences in OS and in PFS between Type 1 and Type 2 or 3 (P < 0.0001 for both), while no difference of OS and PFS was observed between Type 3 and Type 2 (Figure 5). We then combined the results of Type 3 and 2; Type 2/3 predicted extremely poor OS and PFS relative to the Type 1 (P < 0.0001 for both) (Figure 5).
Figure 5.
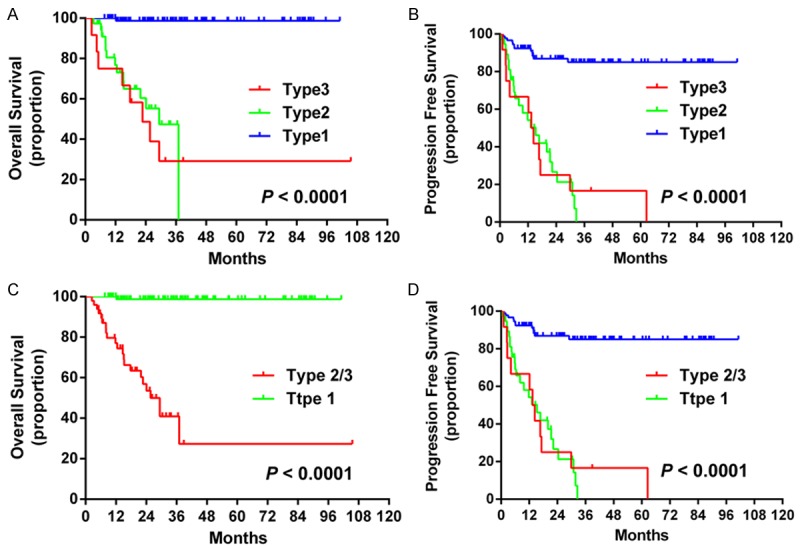
Survival curves for the MA algorithm. The MA algorithm showed significant differences in OS (A) and PFS (B) between patients with Type 1 and 2 or 3. No difference in OS or PFS was observed between those with Type 2 and Type 3 disease. Significant differences in OS (C) and PFS (D) were observed between Type 2/3 and Type 1 groups. Abbreviation: GCB: germinal center B-cells; OS: overall survival; PFS: progression-free survival; MA: MYC aberrations.
Discussion
As a consequence of the work described above, we conclude that a new algorithm, based on MA, which showed a significant prognostic value in DLBCL patients treated with R-CHOP-like therapies. DLBCL is considered aggressive, and predicting the outcome of an individual patient is still difficult. This difficulty stems from the fact that DLBCL is a clinically and biologically heterogeneous group of lymphoma, with no clear histological criteria for subdivision [21]. Although new developments in chemoimmunotherapy, especially the anti-CD20 antibody rituximab, have improved the survival of patients with DLBCL, prognosis prediction is still difficult [22,23]. Currently, GEP, the standard method to designate patients into molecular subsets, is not clinically practical. This fact has led to efforts to find robust, affordable, and reproducible techniques to approximate the information gained from GEP. IHC algorithms that are supposed to be useful surrogates for the classification of DLBCL subsets have been published, most of which use a combination of antibodies against GCB and ABC specific antigens [8,12,15,19]. However, the prognostic power of these algorithms has weakened with the development of new drugs, including rituximab [7,10,11]. Based on previous reports [10,11] and our data, we suggest none of the current algorithms alone dependably predict outcomes for DLBCL patients, especially for those patients receiving an R-CHOP-like therapy.
We then systematically analyzed the prognostic values by examining the predictive value of single markers in the IHC algorithms. It was previously reported that low CD10 expression (< 20% of cells) predicted poor OS in DLBCL patients [24]. However, we didn’t find the correlation between OS/PFS and CD10 expression. Bcl6 is a marker associated with both GCB and ABC subtype [11], which suggests Bcl6 may not a dependable marker used to predict cell of origin [COO] alone. In our study, Bcl6 expression showed no impact on survival. MUM1, as a post- germinal center marker, was once reported to have a negative impact on OS in CHOP-treated patients [25]. Our patients who were treated with R-CHOP-like therapies, however, showed no survival difference on MUM1. We used the two different cut-off values of GCET1 recommended in the Choi and Tally algorithms. Although GCET1 is a marker restricted to GCB subtype [26], neither of the two cut-off values showed different effects on survival. FOXP1, an ABC subtype–associated transcription factor, also seemed to have less prognostic value than when used in the algorithms [27,28], which was confirmed in our study. LMO2, the marker used in the Tally algorithm, was reported to predict an improvement of outcome with or without additional markers [29,30]. In our study, we analyzed the value of LMO2 in the R-CHOP group. LMO2 expression predicted no effect on OS and PFS using a cutoff of 30%. However, in consistency and correlation analyses, LMO2 was found to have a negative relationship with other GCB markers.
Besides the markers included in each algorithm, we also analyzed the proteins beyond those encoded by these algorithms. Myc and Bcl2, for instance, play important roles in predicting prognosis in DLBCL [5,31-33]. As reported in most studies [31,32], Myc expression conferred significantly inferior OS and PFS. Bcl2 expression, however, demonstrated no impact on OS but did contribute to decreased PFS. Since the Bcl2 protein had no consistent prognostic value in previous reports [34,35], more inquiry is still needed to confirm the role of Bcl2 protein in DLBCL.
In addition, we performed FISH tests on MYC and BCL2, which are two classic genes rearranged in double-hit lymphoma [5,6,36,37]. Like Myc expression, MYC rearrangement played a more robust role than BCL2 in predicting the outcome of DLBCL in the R-CHOP-like treatment group. Moreover, we paid great attention to the survival curves, which showed that each algorithm had an obvious survival overlap between GCB and non-GCB subtypes during the initial two years after diagnosis, which means these patients had extremely poor outcomes. A considerable number of these patients had MA (either Myc expression or MYC rearrangement). We therefore incorporated MA into the original algorithms and established a new algorithm (called MA). In accordance with our expectations, patients of MA had much shorter OS and PFS than non-MA. However, the non-MA patients (GCB or non-GCB), still showed no differences in survival (except for PFS of Tally-MA algorithm). One reason for this lack of difference is the heterogeneity of DLBCL, which makes prediction of DLBCL outcomes on the basis of IHC algorithms or prognostic markers alone inexact. The limited number of patients enrolled in our studies might be another factor. Moreover, besides R-CHOP, new targeted drugs have been developed [38,39], which also blurs the survival boundary between GCB and non-GCB.
The MA algorithm could be used to classify DLBCL into three groups (MA, GCB and non-GCB), and our results showed that this division could be used to produce predictions that were better than any other algorithms. Since the final purpose of each algorithm was to predict the different outcome of patients and other algorithms had also used markers not associated with COO [19], we tried to abandon the original algorithms, which mainly depended on COO. Based on this, we established a new algorithm mainly depended on MYC aberrations, which could then classify the patients into three types. The results indeed showed much better power than any of the algorithms published before. Significant differences of OS were observed among patients with the three types of disease, while no difference of survival was seen between Type 2 (either Myc expression or MYC rearrangement) and Type 3 (both Myc expression and MYC rearrangement), mostly because of the limited cases. The Type 2/3 predicted extremely poor survival compared with the Type 1 group. Although not as accurate as GEP, this algorithm relies solely on MYC aberrations, and can be applied in routine practice, and had a better prognostic value than the conventional algorithms. In addition, it was much simpler than other algorithms, most of which applied many antibodies in a compulsory order. We believe that this MA algorithm will be useful in future research to predict outcomes for DLBCL patients. Since we enrolled a relative small group of patients, additional studies are needed to confirm our results and optimize our algorithm.
In summary, our data indicate that IHC algorithms alone are no long sufficient to predict outcomes for DLBCL patients. New prognostic markers may help to distinguish patients with poor survival from the total cohort. We suggested incorporating new markers into the prognostic systems of DLBCL and applying useful detection techniques to improve these systems’ predictive ability.
Acknowledgements
This study was supported by National Natural Science Foundation of China (30971296, 81170485, 81170488, 81370657, 81470328), Key Projects of Health Department of Jiangsu Province (K201108), Jiangsu Province’s Medical Elite Program (RC2011169), National Public Health Grand Research Foundation (201202017), Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institute (JX10231801), Program for Development of Innovative Research Teams in the First Affiliated Hospital of Nanjing Medical University, Project of National Key Clinical Specialty. National Science < Technology Pillar Program (2014BAI09B12), and Project funded by Jiangsu Provincial Special Program of Medical Science (BL2014086). We appreciate the massive efforts by many undergraduate and graduate students, which is the essential foundation for this paper.
Disclosure of conflict of interest
None.
References
- 1.Martelli M, Ferreri AJ, Agostinelli C, Di Rocco A, Pfreundschuh M, Pileri SA. Diffuse large B-cell lymphoma. Crit Rev Oncol Hematol. 2013;87:146–71. doi: 10.1016/j.critrevonc.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, Lefort S, Marit G, Macro M, Sebban C, Belhadj K, Bordessoule D, Ferme C, Tilly H. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood. 2010;116:2040–5. doi: 10.1182/blood-2010-03-276246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Lin P, Young KH, Kanagal-Shamanna R, Yin CC, Medeiros LJ. MYC/BCL2 double-hit high-grade B-cell lymphoma. Adv Anat Pathol. 2013;20:315–26. doi: 10.1097/PAP.0b013e3182a289f2. [DOI] [PubMed] [Google Scholar]
- 6.Horn H, Ziepert M, Becher C, Barth TF, Bernd HW, Feller AC, Klapper W, Hummel M, Stein H, Hansmann ML, Schmelter C, Moller P, Cogliatti S, Pfreundschuh M, Schmitz N, Trumper L, Siebert R, Loeffler M, Rosenwald A, Ott G German High-Grade Non-Hodgkin Lymphoma Study Group. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–63. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 7.Fu K, Weisenburger DD, Choi WW, Perry KD, Smith LM, Shi X, Hans CP, Greiner TC, Bierman PJ, Bociek RG, Armitage JO, Chan WC, Vose JM. Addition of rituximab to standard chemotherapy improves the survival of both the germinal center B-cell-like and non-germinal center B-cell-like subtypes of diffuse large B-cell lymphoma. J. Clin. Oncol. 2008;26:4587–94. doi: 10.1200/JCO.2007.15.9277. [DOI] [PubMed] [Google Scholar]
- 8.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, Tzankov A, Wen W, Liu WM, Kahl BS, d’Amore ES, Montes-Moreno S, Dybkaer K, Chiu A, Tam W, Orazi A, Zu Y, Bhagat G, Winter JN, Wang HY, O’Neill S, Dunphy CH, Hsi ED, Zhao XF, Go RS, Choi WW, Zhou F, Czader M, Tong J, Zhao X, van Krieken JH, Huang Q, Ai W, Etzell J, Ponzoni M, Ferreri AJ, Piris MA, Moller MB, Bueso-Ramos CE, Medeiros LJ, Wu L, Young KH. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–13. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A predictive model for aggressive non-Hodgkin’s lymphoma. The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 10.Hwang HS, Park CS, Yoon DH, Suh C, Huh J. High Concordance of Gene Expression Profiling-correlated Immunohistochemistry Algorithms in Diffuse Large B-cell Lymphoma, Not Otherwise Specified. Am J Surg Pathol. 2014;38:1046–57. doi: 10.1097/PAS.0000000000000211. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho R, Clear AJ, Owen A, Wilson A, Matthews J, Lee A, Alvarez R, Gomes da Silva M, Cabecadas J, Calaminici M, Gribben JG. Poor concordance among nine immunohistochemistry classifiers of cell-of-origin for diffuse large B-cell lymphoma: implications for therapeutic strategies. Clin Cancer Res. 2013;19:6686–95. doi: 10.1158/1078-0432.CCR-13-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 13.Nyman H, Adde M, Karjalainen-Lindsberg ML, Taskinen M, Berglund M, Amini RM, Blomqvist C, Enblad G, Leppa S. Prognostic impact of immunohistochemically defined germinal center phenotype in diffuse large B-cell lymphoma patients treated with immunochemotherapy. Blood. 2007;109:4930–5. doi: 10.1182/blood-2006-09-047068. [DOI] [PubMed] [Google Scholar]
- 14.Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, Connors JM, Gratzinger D, Rosado M, Zhao S, Pohlman B, Wongchaowart N, Bast M, Avigdor A, Schiby G, Nagler A, Byrne GE, Levy R, Gascoyne RD, Lossos IS. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J. Clin. Oncol. 2008;26:447–54. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 15.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, Braziel RM, Geng H, Iqbal J, Lenz G, Vose JM, Hans CP, Fu K, Smith LM, Li M, Liu Z, Gascoyne RD, Rosenwald A, Ott G, Rimsza LM, Campo E, Jaffe ES, Jaye DL, Staudt LM, Chan WC. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia ZG, Xu ZZ, Zhao WL, Zhao SQ, Ding F, Chen Y, Chen QS, Zheng Y, Zhu Q, Hu JP, Shen ZX, Li JM. The prognostic value of immunohistochemical subtyping in Chinese patients with de novo diffuse large B-cell lymphoma undergoing CHOP or R-CHOP treatment. Ann Hematol. 2010;89:171–7. doi: 10.1007/s00277-009-0799-2. [DOI] [PubMed] [Google Scholar]
- 17.Seki R, Ohshima K, Fujisaki T, Uike N, Kawano F, Gondo H, Makino S, Eto T, Moriuchi Y, Taguchi F, Kamimura T, Tsuda H, Ogawa R, Shimoda K, Yamashita K, Suzuki K, Suzushima H, Tsukazaki K, Higuchi M, Utsunomiya A, Iwahashi M, Imamura Y, Tamura K, Suzumiya J, Yoshida M, Abe Y, Matsumoto T, Okamura T. Prognostic impact of immunohistochemical biomarkers in diffuse large B-cell lymphoma in the rituximab era. Cancer Sci. 2009;100:1842–7. doi: 10.1111/j.1349-7006.2009.01268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyman H, Jerkeman M, Karjalainen-Lindsberg ML, Banham AH, Leppa S. Prognostic impact of activated B-cell focused classification in diffuse large B-cell lymphoma patients treated with R-CHOP. Mod Pathol. 2009;22:1094–101. doi: 10.1038/modpathol.2009.73. [DOI] [PubMed] [Google Scholar]
- 19.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, Ott G, Rosenwald A, Braziel RM, Campo E, Vose JM, Lenz G, Staudt LM, Chan WC, Weisenburger DD. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J. Clin. Oncol. 2011;29:200–7. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe ES, Harris NL, Stein H, Isaacson PG. Classification of lymphoid neoplasms: the microscope as a tool for disease discovery. Blood. 2008;112:4384–99. doi: 10.1182/blood-2008-07-077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues. 4th edition. Lyon: IARC; 2008. pp. 233–7. [Google Scholar]
- 22.Coiffier B. Rituximab therapy in malignant lymphoma. Oncogene. 2007;26:3603–13. doi: 10.1038/sj.onc.1210376. [DOI] [PubMed] [Google Scholar]
- 23.Hunt KE, Reichard KK. Diffuse large B-cell lymphoma. Arch Pathol Lab Med. 2008;132:118–24. doi: 10.5858/2008-132-118-DLBL. [DOI] [PubMed] [Google Scholar]
- 24.Ohshima K, Kawasaki C, Muta H, Muta K, Deyev V, Haraoka S, Suzumiya J, Podack ER, Kikuchi M. CD10 and Bcl10 expression in diffuse large B-cell lymphoma: CD10 is a marker of improved prognosis. Histopathology. 2001;39:156–62. doi: 10.1046/j.1365-2559.2001.01196.x. [DOI] [PubMed] [Google Scholar]
- 25.De Mello CA, De Andrade VP, De Lima VC, Carvalho AL, Soares FA. Prognostic impact of MUM1 expression by immunohistochemistry on primary mediastinal large B-cell lymphoma. Leuk Lymphoma. 2011;52:1495–503. doi: 10.3109/10428194.2011.573032. [DOI] [PubMed] [Google Scholar]
- 26.Montes-Moreno S, Roncador G, Maestre L, Martinez N, Sanchez-Verde L, Camacho FI, Cannata J, Martinez-Torrecuadrada JL, Shen Y, Chan WC, Piris MA. Gcet1 (centerin), a highly restricted marker for a subset of germinal center-derived lymphomas. Blood. 2008;111:351–8. doi: 10.1182/blood-2007-06-094151. [DOI] [PubMed] [Google Scholar]
- 27.Wong KK, Gascoyne DM, Brown PJ, Soilleux EJ, Snell C, Chen H, Lyne L, Lawrie CH, Gascoyne RD, Pedersen LM, Moller MB, Pulford K, Murphy D, Green TM, Banham AH. Reciprocal expression of the endocytic protein HIP1R and its repressor FOXP1 predicts outcome in R-CHOP-treated diffuse large B-cell lymphoma patients. Leukemia. 2014;28:362–72. doi: 10.1038/leu.2013.224. [DOI] [PubMed] [Google Scholar]
- 28.He M, Gao L, Zhang S, Tao L, Wang J, Yang J, Zhu M. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL Gastric Cancer. Gastric Cancer. 2014;17:431–41. doi: 10.1007/s10120-013-0313-3. [DOI] [PubMed] [Google Scholar]
- 29.Lossos C, Bayraktar S, Weinzierl E, Younes SF, Hosein PJ, Tibshirani RJ, Sutton Posthumus J, DeAngelis LM, Raizer J, Schiff D, Abrey L, Natkunam Y, Lossos IS. LMO2 and BCL6 are associated with improved survival in primary central nervous system lymphoma. Br J Haematol. 2014;165:640–8. doi: 10.1111/bjh.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alizadeh AA, Gentles AJ, Alencar AJ, Liu CL, Kohrt HE, Houot R, Goldstein MJ, Zhao S, Natkunam Y, Advani RH, Gascoyne RD, Briones J, Tibshirani RJ, Myklebust JH, Plevritis SK, Lossos IS, Levy R. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118:1350–8. doi: 10.1182/blood-2011-03-345272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valera A, Lopez-Guillermo A, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Espinosa I, Novelli S, Briones J, Mate JL, Salamero O, Sancho JM, Arenillas L, Serrano S, Erill N, Martinez D, Castillo P, Rovira J, Martinez A, Campo E, Colomo L Grup per l’Estudi dels Limfomes de Catalunya i Balears (GELCAB) MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica. 2013;98:1554–62. doi: 10.3324/haematol.2013.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, Liu WM, Visco C, Li Y, Miranda RN, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, Zu Y, Bhagat G, Richards KL, Hsi ED, Choi WW, Zhao X, van Krieken JH, Huang Q, Huh J, Ai W, Ponzoni M, Ferreri AJ, Zhou F, Slack GW, Gascoyne RD, Tu M, Variakojis D, Chen W, Go RS, Piris MA, Moller MB, Medeiros LJ, Young KH. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–31. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green TM, Young KH, Visco C, Xu-Monette ZY, Orazi A, Go RS, Nielsen O, Gadeberg OV, Mourits-Andersen T, Frederiksen M, Pedersen LM, Moller MB. Immunohistochemical Double-Hit Score Is a Strong Predictor of Outcome in Patients With Diffuse Large B-Cell Lymphoma Treated With Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone. Journal of Clinical Oncology. 2012;30:3460–7. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 34.Iqbal J, Meyer PN, Smith LM, Johnson NA, Vose JM, Greiner TC, Connors JM, Staudt LM, Rimsza L, Jaffe E, Rosenwald A, Ott G, Delabie J, Campo E, Braziel RM, Cook JR, Tubbs RR, Gascoyne RD, Armitage JO, Weisenburger DD, Chan WC. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin Cancer Res. 2011;17:7785–95. doi: 10.1158/1078-0432.CCR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iqbal J, Neppalli VT, Wright G, Dave BJ, Horsman DE, Rosenwald A, Lynch J, Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Campo E, Ott G, Muller-Hermelink HK, Delabie J, Jaffe ES, Grogan TM, Connors JM, Vose JM, Armitage JO, Staudt LM, Chan WC. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J. Clin. Oncol. 2006;24:961–8. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen MO, Gang AO, Poulsen TS, Knudsen H, Lauritzen AF, Nielsen SL, Gang UO, Norgaard P. Double-hit BCL2/MYC translocations in a consecutive cohort of patients with large B-cell lymphoma. Eur J Haematol. 2012;89:63–71. doi: 10.1111/j.1600-0609.2012.01787.x. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi T, Tsutsumi Y, Sakamoto N, Nagoshi H, Yamamoto-Sugitani M, Shimura Y, Mizutani S, Matsumoto Y, Nishida K, Horiike S, Asano N, Nakamura S, Kuroda J, Taniwaki M. Double-hit lymphomas constitute a highly aggressive subgroup in diffuse large B-cell lymphomas in the era of rituximab. Jpn J Clin Oncol. 2012;42:1035–42. doi: 10.1093/jjco/hys148. [DOI] [PubMed] [Google Scholar]
- 38.Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, Duveau DY, Jiang JK, Michael S, Mierzwa T, Huang W, Walsh MJ, Mott BT, Patel P, Leister W, Maloney DJ, Leclair CA, Rai G, Jadhav A, Peyser BD, Austin CP, Martin SE, Simeonov A, Ferrer M, Staudt LM, Thomas CJ. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014;111:2349–54. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang LH, Kosek J, Wang M, Heise C, Schafer PH, Chopra R. Lenalidomide efficacy in activated B-cell-like subtype diffuse large B-cell lymphoma is dependent upon IRF4 and cereblon expression. Br J Haematol. 2013;160:487–502. doi: 10.1111/bjh.12172. [DOI] [PubMed] [Google Scholar]


