Figure 1. Illustration of codon alignment generation process.
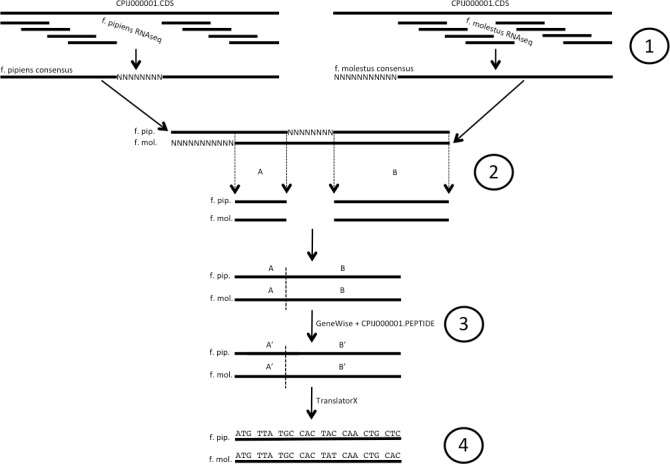
(1) Illumina short read data are aligned to Cx. quinquefasciatus reference CDS sequence and used to build consensus sequences for both Cx. pipiens forms pipiens and molestus. (2) Consensus sequences for each gene are aligned, homologous positions free of Ns are removed and spliced. (3) GeneWise is used along with the corresponding full length Cx. quinq. peptide to create in-frame f. pipiens/f. molestus EST sequences from spliced alignments. (4) Codon alignments are created from EST sequences using TranslatorX. Ns denote unknown and/or unrecovered nucleotide data.
