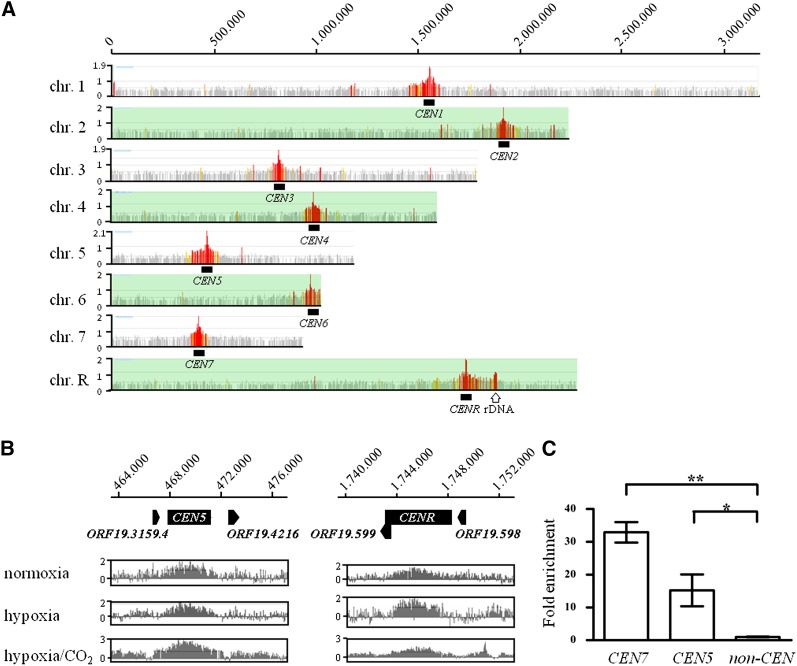
Figure 1.
Genomic localization of the Sch9 kinase. The ChIP-chip procedure was carried out essentially as described previously (Lassak et al. 2011; Schaekel et al. 2013). C. albicans genomic tiling microarrays (NimbleGen) were probed pair-wise by immunoprecipitated chromatin of a strain expressing HA-tagged Sch9 (AF1006) and the corresponding control strain (CAS1). Two independent cultures were assayed for each combination of strains. (A) An overview of Sch9 binding. Significant binding peaks were calculated by the NimbleScan software (NimbleGen) and color-coded according to their FDR values in red [false discovery rate (FDR) ≤ 0.05], orange (FDR ≤ 0.1), yellow (FDR 0.1–0.2), and gray (FDR > 0.2). Significant Sch9-binding peaks were detected at centromeres by genomic ChIP-chip on all C. albicans chromosomes. In addition, a significant peak occurred at the rDNA locus (open arrow). (B) Examples for centromeric binding of Sch9 at CEN5 and CENR. Scaled log2 ratios of cells grown in normoxia and hypoxia with or without 6% CO2 are shown. Note that Sch9 enrichment was obtained for cells grown under normoxia or hypoxia conditions. (C) Enrichment at CEN7, CEN5, and the noncentromeric region was analyzed using qPCR. qPCR analysis reveals the mean fold enrichment of Sch9 at the centromeres obtained in two independent ChIP experiments (±SD) relative to the no-tag control and normalized to the input samples. The calculation was done with two biological replicates (two ChIP samples), and each measurement was performed in triplicate. Significant difference was observed in Sch9 recruitment at the CEN5 and CEN7 region (P < 0.05) and (P < 0.01), respectively (shown by asterisks).