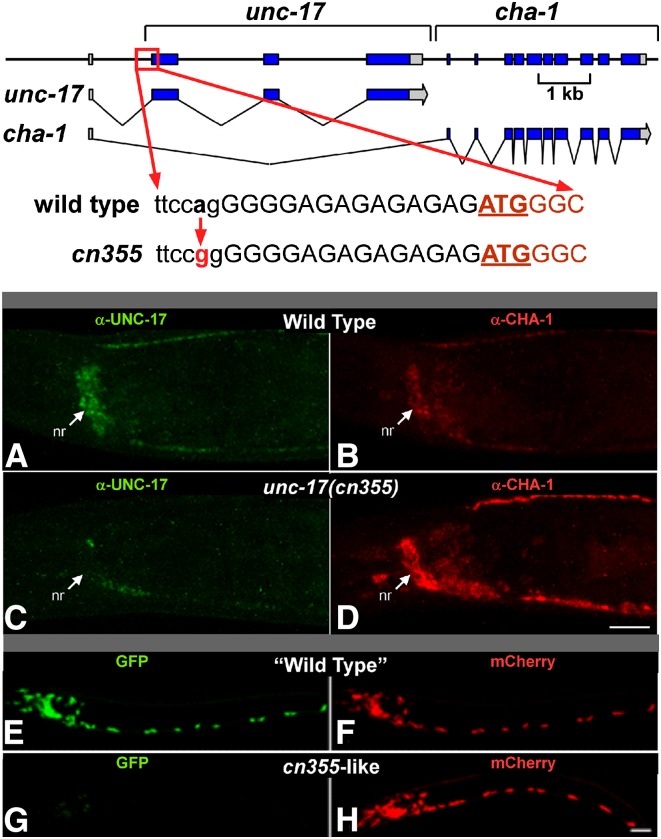
Figure 4.
Validation of the unc-17–cha-1 dual reporter. Top: The unc-17(cn355) a > g mutation disrupts the splice acceptor site (in lowercase) required for production of the endogenous unc-17 transcript; this is predicted to reduce UNC-17 expression. The UNC-17 initiation codon is underlined. There are several weak upstream splice acceptor sequences; it is likely that collectively these account for the low level of residual unc-17 transcripts present in cn355 animals. Middle (A–D): Wild-type (A and B) and cn355 mutant (C and D) animals were double stained under identical conditions with anti-UNC-17 (A and C) and anti-CHA-1 (B and D) antibodies. Shown are head regions of young adults. nr, nerve ring; anterior is to the left, and ventral is down. Bar, 15 µm. Compared to wild type, cn355 homozygotes have reduced UNC-17 expression and increased CHA-1 expression. Bottom (E–H): When introduced into the dual reporter, the cn355 mutation had comparable effects on expression. L1 animals were imaged so that the ratio of red to green fluorescence (R/G) was ∼1.0 with the “wild-type” transgene (E and F). Green is GFP (E and G, corresponding to unc-17) and red is mCherry (F and H, corresponding to cha-1); anterior is to the left, and ventral is down. Bar, 10 µm.