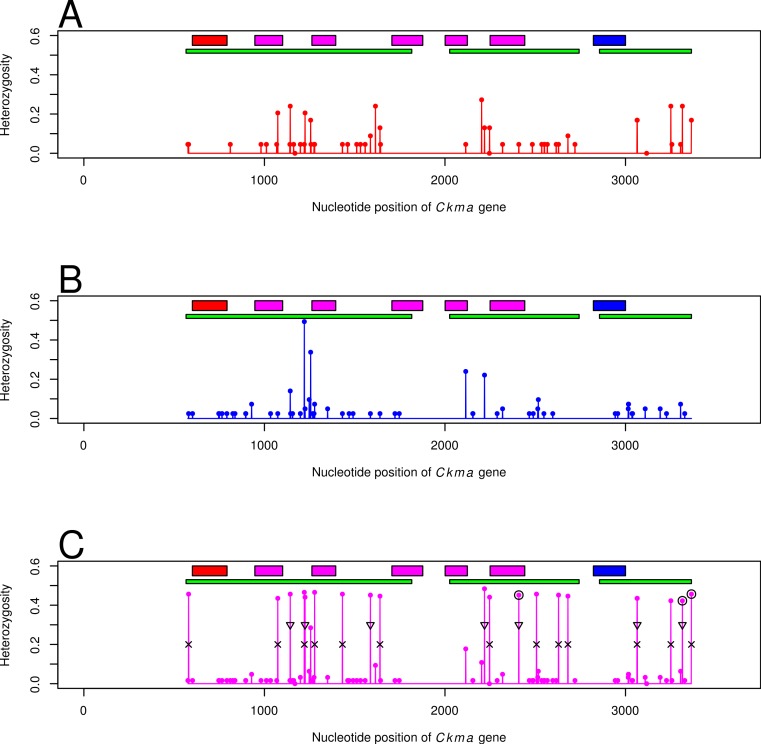
Figure 2. Heterozygosity per nucleotide site of Ckma locus among A alleles (red A, n = 43), B alleles (blue B, n = 79), and all individuals combined (magenta C, n = 122).
Boxes represent exons, start (red), internal (magenta) and terminal (blue). Green boxes represent sequenced fragments trimmed to Phred score of at least 30. The black circles mark the three SNPs of Moen et al. (2008), Gm366-0514 locus with an FST = 0.83, Gm366-1022 locus with an FST = 0.82, and Gm366-1073 with an FST = 0.82 from left to right respectively. Crosses mark mutant sites relative to outgroup that were fixed or nearly fixed among A alleles. Triangles mark mutant sites relative to outgroup that were fixed or nearly fixed among B alleles. Gadus macrocephalus individual 152047 was used as the outgroup.