Dear Editor,
Aptamers are short stretches of nucleotides or amino-acid residues that are engineered to bind to various targets, from small chemicals to large proteins and live cells1,2,3,4,5. Aptamers are normally selected through a process called systematic evolution of ligands by exponential enrichment (SELEX), in which aptamers are selected from a random library of oligonucleotides or peptides against a target through repeated rounds of selection and amplification2. As a versatile affinity reagent, aptamers can be used to modulate the behavior of cells6,7, to detect chemical substances in solution8,9, as alternatives to antibodies5,10 and as therapeutic reagents for human diseases11. As aptamers are chemically synthesized in vitro, they offer a great advantage in selection, production, transportation and storage over traditional antibodies. Also, they can come with many types of chemical modifications, which is useful in cell biology. Although many aptamers have been selected against different tissues and cell types, including mouse embryonic stem cells (mESCs)12, aptamers that specifically label human pluripotent stem cells (PSCs) are still lacking, likely due to the delicate nature of these cells. In this report, we present the first in-depth analysis of a DNA aptamer that specifically labels PSCs. We show that this aptamer can be used as an excellent affinity reagent and functions in hESC enrichment and depletion, and in induced PSC (iPSC) enrichment during reprogramming. We also present a novel approach based on the ratio of epitope expression levels across different cell types to quickly identify binding targets of a given molecule among thousands of proteins on the cell surface, bypassing the need of pull-down experiments and mass spectrometry.
We recently performed a whole-cell SELEX on human PSCs (Supplementary information, Data S1) followed by next-generation sequencing13. Using a novel bioinformatics approach, we identified eight potential aptamers that bind to human PSCs13. Here, we aimed to characterize in detail one of the high-affinity aptamers, Apt19, and to explore its application in stem cell biology.
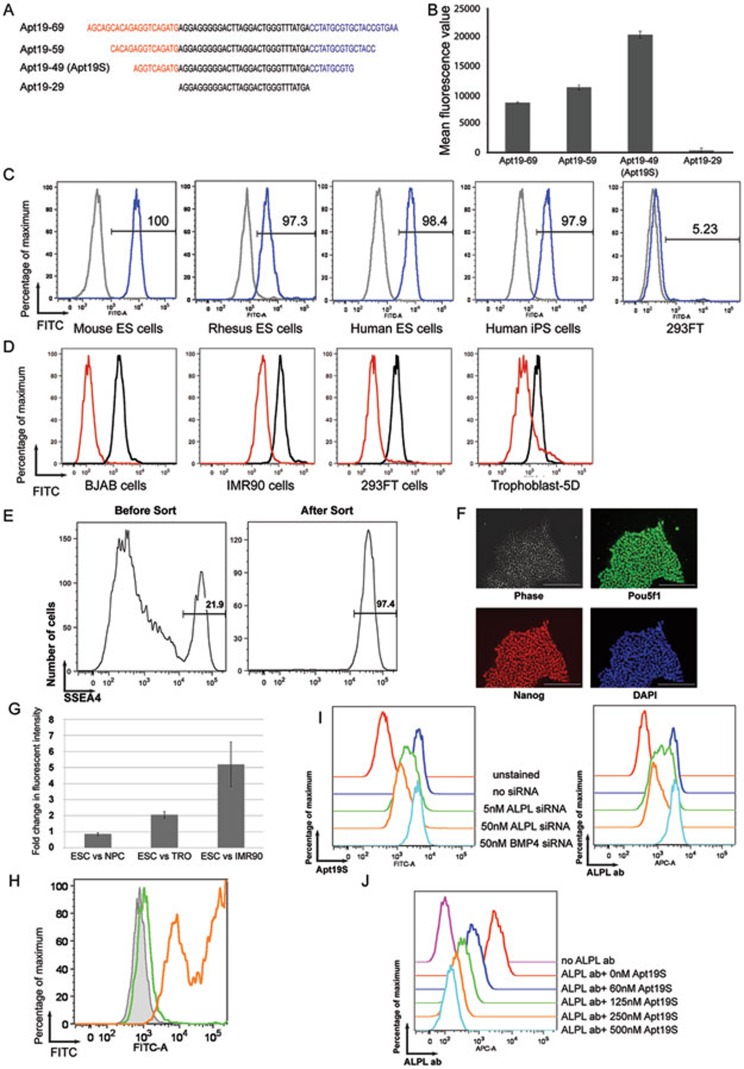
We first examined the contribution of constant regions at either end of the aptamer. Constant regions served as primer-binding sites in the PCR amplification of aptamer libraries during selection. Their contribution to the function of a specific aptamer remained unclear. We performed serial truncations in the constant regions of Apt19 (Figure 1A). As shown in Figure 1B, Apt19's affinity to hESCs was slightly enhanced upon a truncation of 5 nucleotides (nt) at both ends. A truncation of 10 nt at both ends significantly increased Apt19's affinity to hESCs (Figure 1B and Supplementary information, Figure S1A). However, a further truncation of the entire constant region at both ends reduced Apt19's affinity to hESCs to background level (Figure 1B and Supplementary information, Figure S1A). These data suggest that the constant region of an aptamer plays a complicated role in its function. Due to its higher affinity and lower synthesis cost, we used Apt19 with 10-nt truncation at both ends in all of our later studies and named this shorter aptamer Apt19S. We also synthesized a scrambled version of Apt19S (Apt19S-SC) to serve as a negative control, which contains the same 5′ and 3′ truncated constant regions but with a scrambled variable region (Supplementary information, Figure S1B). Circular dichroism analysis showed that Apt19S had a much higher signal at 260 nm compared with Apt19S-SC (Supplementary information, Figure S1C), suggesting the presence of G-quadruplex structure14.
Figure 1.
Characterization and target identification of a DNA aptamer for PSCs. (A) Diagram showing the location of truncations made in the constant regions of Apt19. (B) The affinity of aptamers in A towards hESC was measured with flow cytometry (error bar, SEM; n = 3). (C) Different types of PSCs were incubated with FITC-labeled Apt19S (blue) or Apt19S-SC (grey). The fluorescent intensities of the cells were then measured by flow cytometry. Human 293FT cells were used as a negative control. (D) Different types of differentiated human cell types were incubated with FITC-labeled Apt19S (red) and analyzed by flow cytometry. Human ESCs incubated with Apt19S were used as a positive control (black). (E) A mixture of hESCs and human foreskin fibroblast cells were incubated with biotin-labeled Apt19S and FITC-labeled SSEA4 antibody. After incubation, Apt19S was separated from unbound cells by streptavidin-labeled magnetic beads. The fluorescence profiles of the cell mixture before streptavidin beads separation (left) and cells bound to the beads (right) were analyzed by flow cytometry. The number in each plot shows the percentage of SSEA4+ hESCs in the cell mixture. (F) Beads-bound cells in E were expanded in culture and stained for pluripotency markers Pou5f1 and Nanog. Cell nuclei were labeled with DAPI (scale bar, 200 μm). (G) Fluorescently labeled Apt19S was incubated with H1 ESCs, NPCs, TRO cells and IMR90 fibroblast cells. The fluorescent intensity of each cell type was measured by flow cytometry after incubation and presented as the fold changes between H1 cells and other cell types (error bar, SEM; n = 3). (H) Fluorescently labeled Apt19S was incubated with 293FT cells (grey shade), 293FT cells overexpressing ALPL (orange line) or PROM1 (green line). The fluorescent profiles were then measured by flow cytometry. (I) The binding of fluorescently labeled Apt19S (left panel) or an ALPL antibody (right panel) to hESCs treated with siRNAs against either ALPL or BMP4 was analyzed by flow cytometry. (J) Binding of a fluorescently labeled ALPL antibody to hESCs in the presence of increasing amounts of Apt19S was analyzed by flow cytometry.
We next tested whether Apt19S's affinity was specific to hESCs. We incubated Apt19S with human iPSCs, mESCs and Rhesus monkey ESCs and found that Apt19S showed significant affinity towards all PSCs tested (Figure 1C). We then tested its affinity to non-PSCs. As shown in Figure 1D, Apt19S's affinity to all non-PSCs tested was much lower compared with that to hESCs. Even trophoblast cells differentiated from hESCs for only 5 days showed a significant drop in Apt19S binding (Figure 1D). Truncation of constant regions did not change the specificity of Apt19 as full-length Apt19 showed similar binding profiles towards different cell types (Supplementary information, Figure S1D and S1E). These results suggest that Apt19S binds specifically to epitopes present on PSCs. This property of Apt19S also opens the possibility of using it as a sorting reagent to separate PSCs from non-PSCs. This application is important in regenerative medicine as contamination of undifferentiated PSCs is a major safety concern in the clinical use of cells derived from PSCs. To test the possibility of using Apt19S in cell separation, we incubated a mixture of human foreskin fibroblast (HFF) cells and hESCs with a biotinylated Apt19S. After the incubation, we separated Apt19S-labeled cells from the cell mixture with streptavidin-labeled magnetic beads and analyzed cells that bound to the beads (Figure 1E) and those that remained in the solution (unbound cells) (Supplementary information, Figure S1F). As shown in Figure 1E, we obtained hESCs with a purity of > 97% after a single purification with Apt19S from a cell mixture containing only ∼20% of hESCs. We were also able to reduce hESC percentage by 50-fold in the unbound fraction using Apt19S, whereas Apt19S-SC failed to do so (Supplementary information, Figure S1F). Importantly, hESCs purified using this approach could be further expanded in culture and maintained the expression of pluripotency markers (Figure 1F). We then tested whether Apt19S can be used to enrich iPSCs in reprogramming experiments. As shown in Supplementary information, Figure S1G, we were able to get 6-fold more iPSCs using Apt19S-mediated enrichment. Also, the unbound fraction of Apt19S was almost completely void of any iPSCs (Supplementary information, Figure S1H), indicating highly efficient binding of Apt19S to iPSCs. The enriched iPSCs could also be further expanded and still maintained the expression of pluripotency markers (Supplementary information, Figure S1I).
To further characterize Apt19S, we sought to identify its target on the cell surface. Target identification of aptamers remains a very challenging task. In place of the traditional affinity pull-down experiments, we decided to use a different approach by taking advantage of the fact that Apt19S has different affinity to different cell types. We first compared the binding affinity of fluorescently labeled Apt19S with progenitor cells derived from hESCs, i.e., neural progenitor cells (NPCs) and trophoblast progenitor (TRO) cells, and terminally differentiated cells, IMR90 fibroblasts, with that to hESCs. We assumed that the fluorescent intensity measured by flow cytometry correlates with the amount of epitopes on the cell surface. As shown in Figure 1G and Supplementary information, Figure S1J, NPCs differentiated for 5 days from hESCs had a comparable amount of epitopes on their surfaces. IMR90 cells had roughly 5-fold less epitopes. TRO cells fall between NPCs and IMR90 cells. Using the fold differences in the binding affinity of Apt19S to different cell types that we obtained from the flow cytometry, we filtered a list of membrane proteins according to their expression levels in each cell type tested. The criteria we used to filter the list are as follows: (1) the gene must have a TPM (transcripts per million) > 5 in hESCs to ensure that it is expressed; (2) the gene must have at least a 5-fold higher expression level in hESCs compared with IMR90; (3) the gene must have at least a 3-fold higher expression level in hESC compared with TRO; and (4) the expression level of the gene in NPCs must be within 3-fold of that in hESCs. We then sorted the filtered membrane genes according to their expression levels in hESCs (Supplementary information, Table S1). The highest ranked gene was ALPL, which encodes alkaline phosphatase. To test whether Apt19S targets ALPL, we overexpressed ALPL in 293FT cells, to which Apt19S does not bind (Figure 1C), and tested the binding of Apt19S to these cells afterwards. Indeed, Apt19S showed substantially increased affinity towards 293FT cells overexpressing ALPL (Figure 1H). This was not due to artifacts of overexpressing membrane proteins as overexpression of another membrane protein, PROM1, did not change the binding affinity of Apt19S to 293FT cells (Figure 1H). To further confirm our finding, we tested whether ALPL knockdown by siRNA would decrease Apt19S's affinity to hESCs. As expected, knockdown of ALPL decreased Apt19S's affinity to hESCs (Figure 1I). The decreased level of the binding affinity correlated with that of ALPL on the cell surface as reflected by ALPL staining (Figure 1I). This decrease was also specific as siRNA targeting BMP4 did not affect Apt19S binding (Figure 1I). However, as ALPL is an enzyme, it is possible that Apt19S binds to its products, not the protein itself. To exclude this possibility, we performed an antibody competition assay. We mixed the anti-ALPL antibody with varying amounts of Apt19S and measured the binding of the antibody to hESCs. As shown in Figure 1J, increasing amounts of Apt19S decreased the antibody's binding to hESCs, suggesting a direct binding between Apt19S and ALPL.
The successful identification of ALPL as the target of Apt19S also validates our novel approach for identifying cell surface targets of a given molecule based on epitope expression ratios across cell types. Affinity pull-down followed by mass spectrometry is a robust and the most widely used approach to identify targets of aptamers. However, this approach can be challenging to labs not specialized in biochemistry. First, most targets of aptamers are cell surface proteins. These proteins are notoriously hard to work with due to their high hydrophobicity and low solubility. This often leads to failed pull-down experiments. Second, the amount of proteins obtained in each pull-down experiment is often very low, which makes accurate protein identification through mass spectrometry challenging. In contrast, our approach does not involve protein purification. Based on differential binding affinities of an aptamer to different cell types and the expression profiles of respective genes, we can quickly narrow down the list of candidate targets. The candidates are then evaluated using binding assays. With the increasing number of whole-transcriptome data sets available in the public and the much lowered cost of RNA sequencing, the transcriptome of a given cell type can be easily obtained, which makes our approach economically very favorable. This approach is also not limited to target identification of aptamers; the cell surface target of any biomolecule (growth factors, antibodies, etc.) can be identified in this way. Nevetheless, our approach might not be able to accurately identify the cell surface targets in certain situations. For instance, when the target is not a protein but instead is a posttranslational modification of a protein or small molecules on the cell surface or when mRNA expression level of a certain gene does not reflect its protein level on the cell surface, the new approach will not be able to identify the target so the traditional pull-down approach should be used.
In summary, we report here the first well-characterized DNA aptamer that specifically labels PSCs. We showed that this aptamer performs as an excellent affinity reagent in both immunostaining and cell separation. Using a novel approach, we also identified ALPL as a target of the aptamer, which is a well-characterized marker for PSCs. However, we cannot rule out the possibility that the aptamer binds cooperatively to another epitope besides ALPL. Our work offers the researchers in the stem cell field a versatile tool and would also expand the use of aptamers in cell biology.
Conflict of interest
JAT is a founder, stockowner, consultant and board member of Cellular Dynamics International (CDI).
Acknowledgments
We thank Mackenzie Holland for editorial assistance. This work was founded by the Morgridge Institute for Research, US National Institutes of Health (1U01ES017166-01 to JAT), and the Foundation for Fighting Blindness (TA-CBT-0607-0444-UWI-WG to JAT).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Flow cytometry histogram showing the binding of Apt19 with different truncations to human ES cells.
Candidate gene identification using whole transcriptome comparison.
Material and methods
References
- Ellington AD, Szostak JW. Nature. 1990. pp. 818–822. [DOI] [PubMed]
- Tuerk C, Gold L. Science. 1990. pp. 505–510. [DOI] [PubMed]
- Gold L, Polisky B, Uhlenbeck O, et al. Annu Rev Biochem. 1995. pp. 763–797. [DOI] [PubMed]
- Cibiel A, Dupont DM, Ducongé F. Pharmaceuticals. 2011. pp. 1216–1235.
- Meyer S, Maufort JP, Nie J, et al. PLoS One. 2013. p. e71798. [DOI] [PMC free article] [PubMed]
- Hicke BJ, Watson SR, Koenig A, et al. J Clin Invest. 1996. pp. 2688–2692. [DOI] [PMC free article] [PubMed]
- Jeong S, Eom T, Kim S, et al. Biochem Biophys Res Commun. 2001. pp. 237–243. [DOI] [PubMed]
- Swensen JS, Xiao Y, Ferguson BS, et al. J Am Chem Soc. 2009. pp. 4262–4266. [DOI] [PMC free article] [PubMed]
- Ferguson BS, Hoggarth DA, Maliniak D, et al. Sci Transl Med. 2013. p. 213ra165. [DOI] [PMC free article] [PubMed]
- Davis KA, Lin Y, Abrams B, et al. Nucleic Acids Res. 1998. pp. 3915–3924. [DOI] [PMC free article] [PubMed]
- Keefe AD, Pai S, Ellington A. Nat Rev Drug Discov. 2010. pp. 537–550. [DOI] [PMC free article] [PubMed]
- Iwagawa T, Ohuchi SP, Watanabe S, et al. Biochimie. 2012. pp. 250–257. [DOI] [PubMed]
- Jiang P, Meyer S, Hou Z, et al. Bioinformatics. 2014. pp. 2665–2667. [DOI] [PMC free article] [PubMed]
- Masiero S, Trotta R, Pieraccini S, et al. Org Biomol Chem. 2010. pp. 2683–2692. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Flow cytometry histogram showing the binding of Apt19 with different truncations to human ES cells.
Candidate gene identification using whole transcriptome comparison.
Material and methods