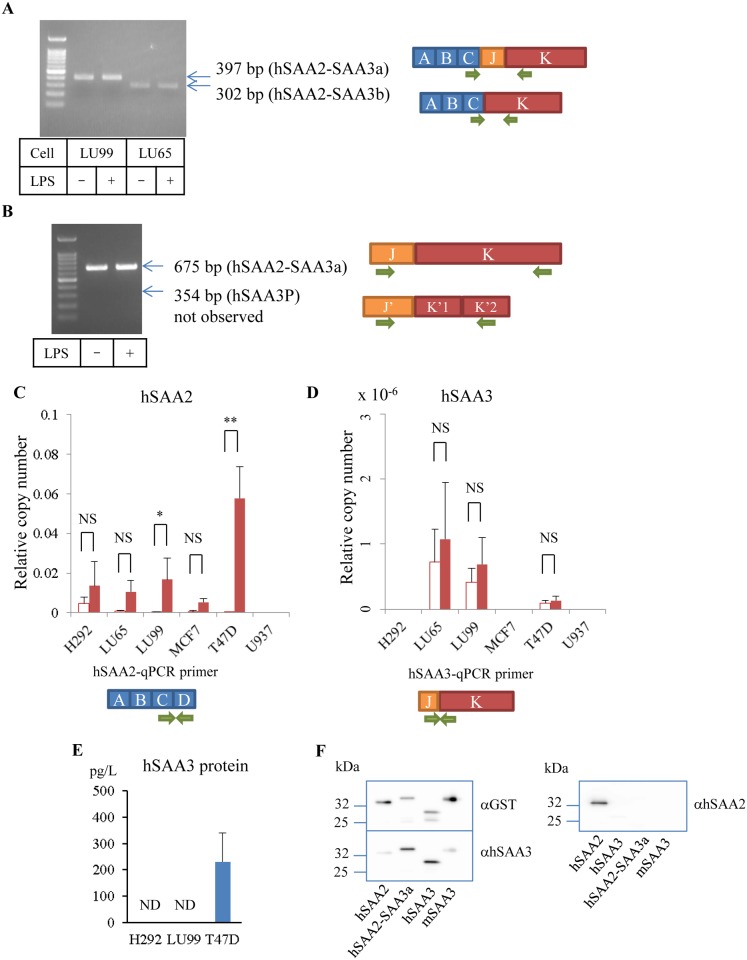
Fig 2. PCR detection of hSAA2-SAA3 readthroughs.
(A) PCR with the hSAA2-SAA3-specific primer set. (B) PCR with the hSAA3-specific primer set. Complimentary DNAs were synthesized using a gene-specific primer that binds hSAA3 exon 2. The PCR products were subjected to DNA sequencing analyses to confirm that these bands were hSAA2-SAA3 readthrough origin (three independent experiments). (C) hSAA2 expression levels normalized to β-actin. *P = 0.0096, **P = 0.0001. (D) hSAA3 expression levels normalized to β-actin. Open bars and filled bars represent without/with IL-1β-IL-6-dexamethazone stimulation, respectively (four independent experiments). NS = not significant (P > 0.05). (E) hSAA3 concentrations determined by Immunoprecipitation (IP)-ELISA. Culture media were passed through the immunoaffiinity column, and the effluents were assayed (four independent experiments). ND = not detected. (F) Western blot analysis of hSAA3 antibody. Purified hSAA2, hSAA2-SAA3a, hSAA3, and mSAA3 proteins fused to GST were blotted onto a membrane and detected with GST (left top), hSAA3 (left bottom), and hSAA2 (right) antibodies.