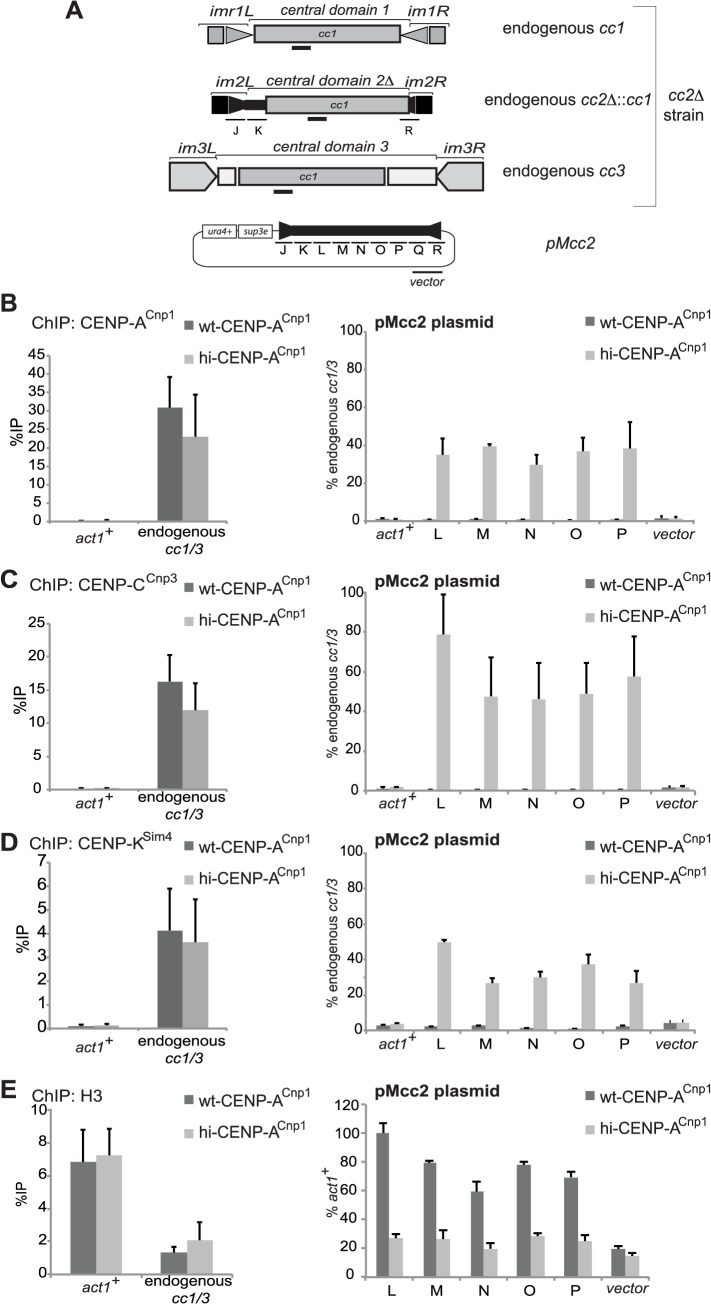
Fig 1. Elevated levels of CENP-ACnp1 are sufficient to establish centromeric chromatin in the absence of heterochromatin.
(A) Schematic representation of the three endogenous centromeres in cc2Δ strains and the plasmid (pMcc2) used in the study. In cc2Δ strains, part of the central core 2 (regions L to Q, each 1 kb) is replaced by 5.5 kb of DNA from central core 1, allowing the analysis of the L-Q sequence within pMcc2. Primer pairs used for quantification at endogenous centromeres recognise the region of homology shared between cc1 and cc3. The product amplified by qPCR for the endogenous centromeres is represented as a black bar. (B) ChIP analysis of CENP-ACnp1 levels at endogenous centromere (cc1/3), act1 +, plasmid backbone (vector) and at cc2 on the plasmid (pMcc2: L-P) in wild-type (wt-CENP-ACnp1) or in the presence of high levels of CENP-ACnp1 (hi-CENP-ACnp1). (C) ChIP analysis for the kinetochore protein CENP-CCnp3. (D) ChIP analysis for the kinetochore protein CENP-KSim4. (E) ChIP analysis of histone H3 levels. For the kinetochore proteins analysed, ChIPs are reported as %IP for endogenous centromeres or as relative to cc1/3 for the pMcc2 plasmid. For the levels of H3, the ChIP analyses are reported as %IP for cc1/3 and as relative to act1 + for the pMcc2 plasmid (n = 3).