Figure 6.
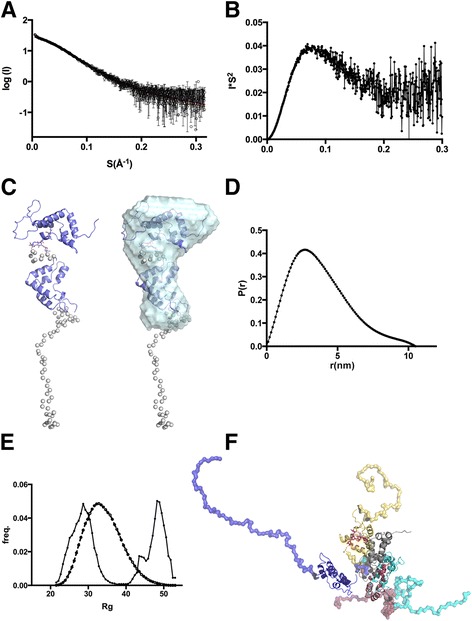
Rigid body and ab initio modelling of GAS1 based on SAXS data. A) Scattering curve and fit of CORAL rigid body model (red line) to observed data. B) The Kratky-plot from the experimental data, suggesting a folded structure with some flexibility. C) CORAL generated model with N- and C-terminal domains as rigid bodies (blue) with flexible linker regions (grey beads; left), and the ab initio model for GAS1 generated by DAMMIN (green; average of 10 calculations) fitted over the rigid body model (right). D) The distance distribution calculated for GAS1 SAXS data. E) SAXS ensemble modelling of GAS1 solution conformations shown as the statistical distribution of Rg-values of best fitted models (continuous line with closed circles) vs. initial random pool (dashed line with open circles) shows a bimodal distribution of GAS1 solution conformations. F) The selected pdb-files representing the ensemble with Chi2 = 0.84 fit to the experimental data, showing extended (blue) and more collapsed models (yellow, red, cyan) in the final ensemble; the N-terminal domain (in grey) was fixed relative to the rest of the protein during the runs. The modelled glycan structure is shown as red “stick” presentation on the N-terminal domain.
