Figure 1.
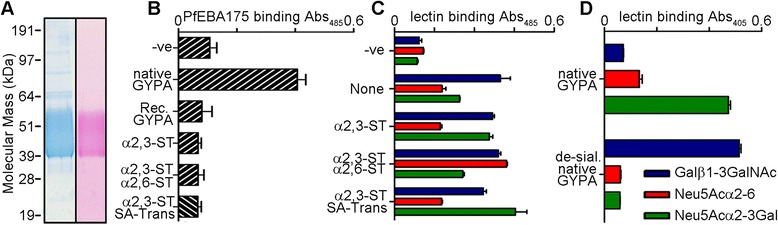
Recombinant soluble GYPA expressed in HEK293E cells is undersialylated and does not bind PfEBA175. (A) Denaturing SDS-PAGE gels of recombinant soluble GYPA, visualized using Coomassie G250 (left panel) and a glycoprotein-specific periodic acid-Schiff reagent stain (right panel). (B) Recombinant soluble biotinylated GYPA co-expressed without (Rec. GYPA) or with plasmids encoding α-2,3 sialyltransferase 1 (α2,3-ST) singly or in combination with an α-2,6 sialyltransferase 1 (α2,6-ST) or a CMP-sialic acid transporter (SA-Trans) were immobilized on a streptavidin-coated plate and tested for their ability to bind pentameric β-lactamase-tagged PfEBA175. The positive control was biotinylated native GYPA and the negative control (−ve) was biotinylated Cd4d3 + 4 alone. (C) Binding of pentameric β-lactamase-tagged recombinant GYPA and its sialylation-enhanced forms by the three lectins that recognize terminal sialic acids (Neu5Ac) or a subterminal Galβ1-3GalNAc disaccharide. Negative control was soluble pentameric β-lactamase-tagged Cd4d3 + 4. The assay was a modified version of AVEXIS with the biotinylated lectins immobilized on a streptavidin-coated plate. (D) Lectin binding to native GYPA and its desialylated (neuraminidase-treated) form to show extent of sialylation of native GYPA. Figure 1D is reproduced from Wanaguru et al. with minor stylistic changes [8]. Bars represent mean ± standard deviation (SD), n = 3.
