Abstract
Background
The ability to increase cellulose content and improve the stem strength of cereals could have beneficial applications in stem lodging and producing crops with higher cellulose content for biofuel feedstocks. Here, such potential is explored in the commercially important crop barley through the manipulation of cellulose synthase genes (CesA).
Results
Barley plants transformed with primary cell wall (PCW) and secondary cell wall (SCW) barley cellulose synthase (HvCesA) cDNAs driven by the CaMV 35S promoter, were analysed for growth and morphology, transcript levels, cellulose content, stem strength, tissue morphology and crystalline cellulose distribution. Transcript levels of the PCW HvCesA transgenes were much lower than expected and silencing of both the endogenous CesA genes and introduced transgenes was often observed. These plants showed no aberrant phenotypes. Although attempts to over-express the SCW HvCesA genes also resulted in silencing of the transgenes and endogenous SCW HvCesA genes, aberrant phenotypes were sometimes observed. These included brittle nodes and, with the 35S:HvCesA4 construct, a more severe dwarfing phenotype, where xylem cells were irregular in shape and partially collapsed. Reductions in cellulose content were also observed in the dwarf plants and transmission electron microscopy showed a significant decrease in cell wall thickness. However, there were no increases in overall crystalline cellulose content or stem strength in the CesA over-expression transgenic plants, despite the use of a powerful constitutive promoter.
Conclusions
The results indicate that the cellulose biosynthetic pathway is tightly regulated, that individual CesA proteins may play different roles in the synthase complex, and that the sensitivity to CesA gene manipulation observed here suggests that in planta engineering of cellulose levels is likely to require more sophisticated strategies.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0448-y) contains supplementary material, which is available to authorized users.
Keywords: Barley, CaMV 35S constitutive promoter, Cellulose, Gene silencing, HvCesA genes, Primary cell walls, Secondary cell walls
Background
In barley, it is estimated that plant lodging can cause a reduction of up to 65% in grain yield [1]. Weakness in the stem and poor root anchorage, when subjected to external factors such as wind, rain or disease, result in stem/root lodging or the permanent failure of the plant shoot to support its upright position [2]. Stem strength is a complex trait reflecting cellulose content, the length, number and arrangement of vascular bundle fibres in the organ, the orientation of cellulose microfibrils and the degree of lignification [3-5]. These traits contribute synergistically to plant stem strength. Previous studies have shown that a decrease in load-bearing cell wall polymers such as cellulose or lignin can negatively affect stem strength in barley [6], wheat [7], rice [8] and maize [9]. In wheat, a combination of Fourier transform infrared resonance (FTIR) analysis, histology and principle component analysis (PCA), showed that cellulose contributed more to stem strength than lignin [10]. Similarly in maize, Appenzeller et al. [11] and Ching et al. [9] showed a strong correlation (r2 = 0.85) between cellulose content (g/cm) and internodal flexural stem strength, but found no consistent correlation between lignin content and stem strength.
Cellulose content therefore seems to be an important contributing factor in stem strength of cereal species. At the molecular level, cellulose consists of linear, unbranched chains of glucosyl residues linked by (1,4)-β-glucosidic linkages [12]. Cellulose chains are often described as flat ribbons that aggregate into microfibrils of 2 to 2.5 nm in thickness. There is some debate as to the precise number of chains that constitute a microfibril, with values ranging from 36 individual (1,4)-β-glucan chains [13] to as few as 16 chains [14]. The microfibrils can further aggregate to form larger macrofibrils and can serve as a scaffold for the non-covalent cross-linking of other non-cellulosic polysaccharides. In primary cell walls, cellulose microfibrils are generally arranged perpendicular to the axis of cell elongation, although the alignment between microfibrils is not strictly parallel. Such an arrangement of microfibrils provides both strength and flexibility that enable the primary cell walls to withstand turgor pressure and to assist in the cell’s directional growth. In the secondary wall, the microfibrils are more organised and are often aligned in parallel arrays. There can be several layers in secondary walls and within each layer the parallel microfibrils can be oriented at different angles to create laminated layers that further strengthen the wall and restrict the cell’s lateral or radial growth.
Data from transcript analyses in barley are consistent with Arabidopsis mutational studies, insofar as the abundance of CesA transcripts in various tissues at different stages of cell wall development, together with co-expression analyses, suggest that two groups of three CesA genes are co-ordinately expressed during the growth of the primary cell wall (PCW) and the secondary cell wall (SCW). In barley, HvCesA1, HvCesA2 and HvCesA6 are believed to be involved in cellulose synthesis during primary cell wall deposition, while HvCesA4, HvCesA7 and HvCesA8 are postulated to participate in cellulose synthesis during SCW deposition; a total of eight HvCesA genes have been identified [15]. It should be noted that these conclusions are based on co-expression of the two groups of three genes and their relatively high transcript levels in tissues that are believed to be undergoing predominantly PCW or SCW deposition. There is no direct evidence in barley that the groups of three enzymes encoded by the three HvCesA genes form a multi-enzyme complex, although this seems likely based on data from other systems [16-20].
In the work described here, barley has been transformed with HvCesA genes driven by the powerful constitutive CaMV 35S promoter, with a view to increasing cellulose content in the walls of transgenic lines and to evaluating the effects of increased cellulose on stem strength. All three PCW HvCesA and two SCW HvCesA genes were studied. The HvCesA5/7 genes were omitted because they appeared to encode enzymes with identical amino acid sequences. The results provide information on the potential for altering cell wall composition in important crop species of the Triticeae from which residual straw, bran from flour milling and spent grain from the brewery might be used in renewable biofuel production.
Results
HvCesA genes are distributed across the grass genome
At least eight barley (Hordeum vulgare) HvCesA genes were identified by Burton and co-authors [15]. With the recent release of the barley scaffold [21,22], a total of nine barley HvCesAs genes has now been identified. In silico mapping of HvCesA genes in barley and two other economically important grasses, Sorghum bicolor (sorghum) and Oryza sativa (rice) indicated that the CesA genes are broadly distributed across the genomes, especially so in barley where HvCesA genes are found on every chromosome except chromosome 4. Figure 1 shows homologous relationships of the CesA genes in barley, sorghum and rice.
Figure 1.
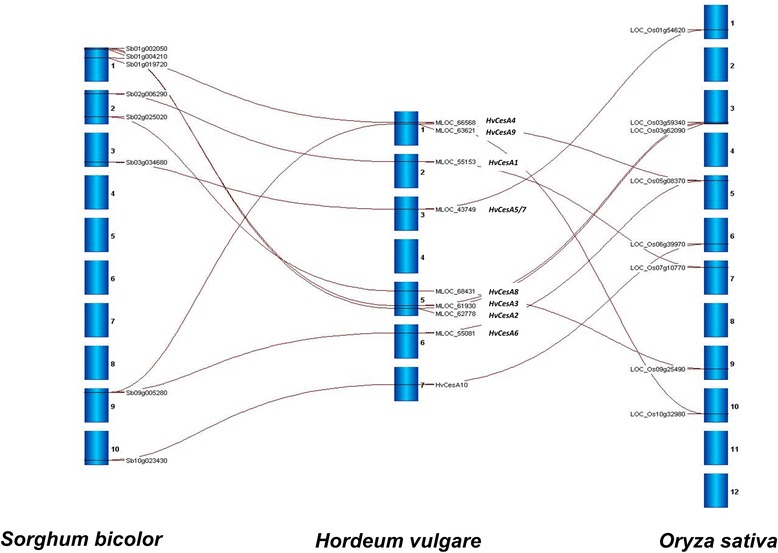
Image generated using Strudel. Gray lines show homologous relationships between CesA genes in Sorghum (Sorghum bicolor), barley (Hordeum vulgare) and rice (Oryza sativa). Positions of CesA genes on the respective chromosomes are also indicated.
Only plants containing SCW 35S:HvCesA constructs exhibit aberrant phenotypes
A total of five constructs driven by the CaMV 35S constitutive promoter were individually transformed into barley. These included the three PCW cellulose synthase cDNAs HvCesA1, HvCesA2 and HvCesA6, and the two SCW cellulose synthase cDNAs HvCesA4 and HvCesA8. Between 13 and 22 transgenic plants per construct were generated.
Most plants (~90%) transformed with PCW HvCesA cDNAs showed no visual abnormalities compared with control Golden Promise barley plants grown under the same conditions. In contrast, more dramatic phenotypes were observed in transgenic plants carrying the SCW HvCesA cDNAs. Dwarfism and early-stage leaf necrosis observed in T035S:HvCesA4 plants persisted into the T1 (Figure 2A) and T2 generations (Figure 2B). At about one month old, these plants were stunted and necrosis was noticeable at leaf tips (Figure 2C). Dwarf plants took a month longer to reach maturity compared with controls. In the T2 generation, about 25% of transgenic progeny from T1 dwarf parents either died or were sterile (Figure 2B), suggesting that this severe phenotype might be linked to a homozygous state for the 35S:HvCesA4 gene. This could not be directly tested but surviving plants showed evidence of segregation; these plants yielded few viable grains.
Figure 2.
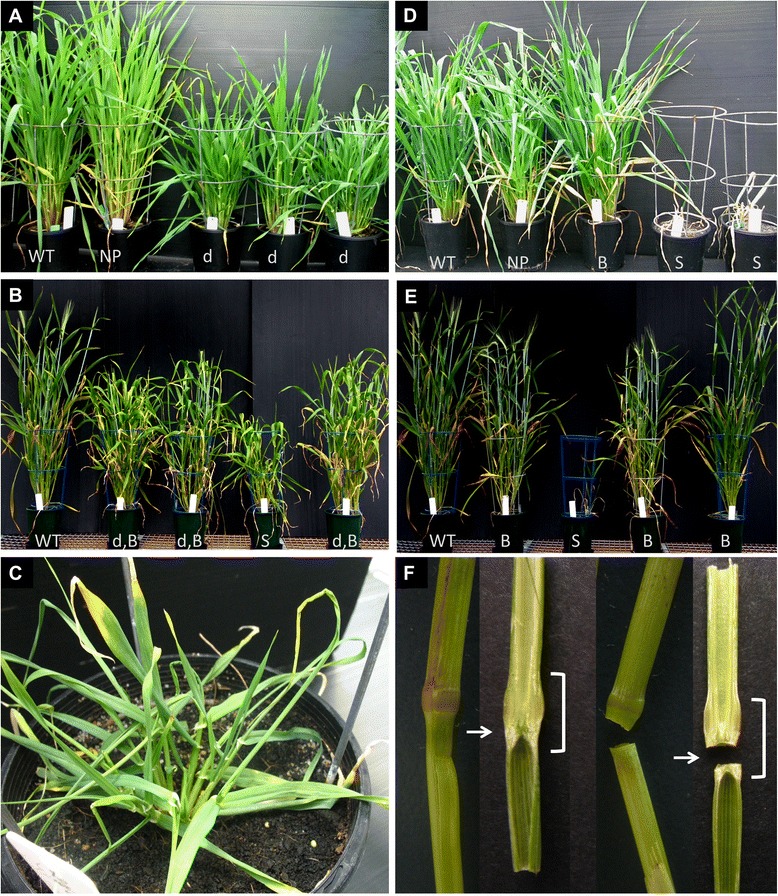
Photos of representatives from the T 1 and T 2 generations showing the aberrant phenotypes observed in 35S:HvCesA4 (A, B,C) and 35S:HvCesA8 (D,E,F) plants. (A) T1 35S:HvCesA4 plants and wild-type (WT) Golden promise on the far left. Dwarfism (d) persisted in most plants grown from parents with an aberrant phenotype except for one or two plants within the same line (e.g. plant NP, normal phenotype). The ratio of plants displaying dwarf: normal phenotype (including nulls & revertants) in T1 is 58%: 42%. (B) Many T2, 35S:HvCesA4 progeny were dwarfed with “brittle nodes” (d,B). About 25% of T2 plants from each line exhibited a severe reduction in stature, was sterile (S) and some died. The plants with a severe phenotype may be homozygotes. (C) Close up view of necrosis found at the leaf-tips of a 1 month old plant that further developed into a dwarf plant with few viable grains. (D) T1 35S:HvCesA8 plants. Aberrant phenotypes observed were “brittle node” (B) and severely stunted plants that died young (S) (~1 month old). Plants with a “brittle node” phenotype had no reduction in stature but when pressure was applied manually, the stems snapped at the nodes. (E) T2 35S:HvCesA8 plants. About 25% of T2 plants from each line were stunted and died young (S). Many were only brittle at the node (B) with no compromise in stature. (F) Comparison of two wild-type (left) and two transgenic “brittle node” stems (right). One stem each from wild-type and transgenic plant were sliced in half to reveal the stem’s internal anatomy. The bracket indicates the nodal region of the stem and at closer inspection the break-point was often found to be at the “nodal plate” (arrow).
For the 35S:HvCesA8 T1 and T2 generations, most plants showed no obvious difference in height, although the putative homozygotes did not grow past the tillering stage (Zadoks’ scale 22) [23] and subsequently died (Figure 2D and Figure 2E). This is similar to the putative homozygotes found in T235S:HvCesA4 plants that were stunted and died at an early stage.
Another feature observed in the T1 and T2 generations of both the 35S:HvCesA4 and 35S:HvCesA8 transgenic plants was brittleness at stem nodes at the heading stage (Figure 2F). This phenotype was apparent for 35S:HvCesA8 in every generation but only became obvious in later generations of 35S:HvCesA4, especially in the T2 generation. The break-point of the brittle node phenotype was usually close to the nodal plate but not found within the stem internode as indicated by a horizontal arrow in Figure 2F. There was also a 45% (3.7 mm down to 2.0 mm) reduction in the average stem diameter of dwarf T135S:HvCesA4 tillers, although no significant difference in the diameter of brittle node T135S:HvCesA8 stems compared with the controls was observed.
Transcript profiles of T0 plants carrying PCW and SCW 35S:HvCesA constructs
The T0 generation of transgenic plants were profiled to determine the effect of PCW and SCW HvCesA manipulations. Transcript profiles were generated for sets of transgenic plants carrying the three PCW HvCesA cDNAs, namely 35S:HvCesA1, 35S:HvCesA2 and 35S:HvCesA6 and for the two SCW 35S:HvCesA4 and 35S:HvCesA8 cDNAs. For each set of HvCesA transgenic plants, transcript levels for the corresponding endogenous genes were also examined (designated eHvCesA1, eHvCesA2 and eHvCesA6, eHvCesA4 and eHvCesA8 [15]). Primers for these endogenous genes are selective and do not amplify the transgene transcript.
Transcript levels for all PCW HvCesA transgenes were low with less than 10% of the levels of transcripts for the corresponding eHvCesA genes expressed in control plants (Additional file 1: Figures S1A cf. B, S2A cf. C, S3A cf. D and S4A cf.B, C cf. D). Although varying levels of transcript were observed for eHvCesA1, eHvCesA2 and eHvCesA6 genes in the three transgenic plant sets, the transcript levels for the endogenous genes were generally lower or equal to those measured in control plants (Additional file 1: Figures S1B-D cf. S2B-D cf. S3B-D and Additional file 1: Table S1). This indicated that both the transgene and endogenous PCW HvCesA gene transcript levels were often suppressed in the transgenic lines.
Similarly, the transgene and its endogenous counterpart were profiled in the SCW HvCesA T0 generation and suppression of endogenous SCW genes was also observed for both constructs (Additional file 1: Figures S4-S5). Unlike PCW transgenic sets, a higher up-regulation of SCW CesA transgene levels was observed and measured to be more than 10% of the endogenous levels in control plants. The HvCesA8 transgene achieved the highest level of up-regulation, measuring 60% of transgene/endogenous ratio (Additional file 1: Table S1).
Transcript profiles in T1 plants containing SCW 35S:HvCesA constructs
Only subsequent generations of plants carrying SCW constructs were studied, because they exhibited less transgene suppression and hence more likely to have an increased cellulose content. In addition, the observation of drastically distinct phenotypes between the SCW transgenic plants allowed comparisons between the 35S:HvCesA4 and 35S:HvCesA8 constructs.
Transcript profiles for both SCW transgenic plants showed plants with either aberrant or normal phenotypes, with each phenotype described represented by three independent segregating lines (Figures 3A and 3B). A striking similarity was observed between plants of T135S:HvCesA4 and T135S:HvCesA8, where suppression of endogenous transcript was accompanied by dwarfism. In line with a more severe gene suppression in T135S:HvCesA8 lines, these plants died early but those in T135S:HvCesA4 survived to maturity. Dwarfed plants had a lower level of endogenous transcript relative to control and normal-looking plants. Normal plants in T135S:HvCesA4 showed a two-fold transgene up-regulation and maintained endogenous transcript levels similar to those in control plants (Table 1). However, normal plants in T135S:HvCesA8 had lower endogenous transcript levels relative to control plants, probably to compensate for the five-fold increase of transgene. Detailed transcript profiles for the transgene and the endogenous eHvCesA4, eHvCesA7 and eHvCesA8 genes for T1 SCW plants relative to control plants are shown in Table 1.
Figure 3.
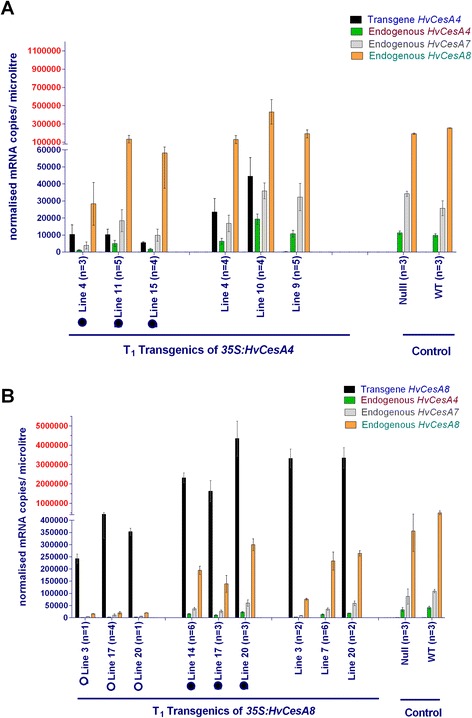
Averaged transcript levels of four genes in transgenic 35S:HvCesA4 and 35S:HvCes8 T 1 plants. X-axis depicts the transgenic lines and control plants (where n = number analysed). The transcript values were averaged for sibling lines with similar phenotype. Where possible, null segregants were selected from three different parental lines. For clarity between very high and low transcript levels, the Y-axis for normalised mRNA copies/microlitre is divided into two different scales (black and red). Error bar is the standard error of the mean (SEM) of biological variation between sibling lines. (A) Transcripts measured for SCW 35S:HvCesA4 transgenic plants were the HvCesA4 transgene and eHvCesA4, eHvCesA7 and eHvCesA8. Plants within the same line exhibited variations in phenotype. There were three independent lines with a dwarfed phenotype (black solid circle) and three other with a normal phenotype. (B) Transcripts measured for SCW 35S:HvCesA8 transgenic plants were the HvCesA8 transgene and eHvCesA4,eHvCesA7 and eHvCesA8. There were three lines that were stunted, sterile and died young (open circle), three lines with a “brittle node” phenotype (black solid circle) and three lines with a normal phenotype.
Table 1.
Percentage gene levels in transgenic vs control plants
| Transgenic T 1 35S:HvCesA4 | Transgene/Endogenous ratio | Endogenous HvCesA4 | Endogenous HvCesA7 | Endogenous HvCesA8 |
| dwarf | 82% | 25% | 36% | 32% |
| normal | 215% | 115% | 95% | 112% |
| Transgenic T 1 35S:HvCesA8 | Transgene/Endogenous ratio | Endogenous HvCesA4 | Endogenous HvCesA7 | Endogenous HvCesA8 |
| stunted | 79% | 6% | 7% | 4% |
| brittle node | 644% | 44% | 42% | 14% |
| normal | 518% | 32% | 35% | 44% |
Values are calculated as [(T/E)*100] to determine the ratio of transgene transcript levels to its corresponding endogenous gene expressed in control plants, where T = Average gene levels in transgenic plants and E = Average corresponding endogenous gene level in control plants. For endogenous genes, percentage expression in transgenic cf. control plants were calculated.
Another observation in T135S:HvCesA4 was that, regardless of the phenotypes observed, tight co-regulation between the three endogenous genes was maintained across the whole transgenic set (correlation coefficients, r2, of 0.85 to 0.99), indicating that the dwarf phenotype did not perturb the coordination of gene transcription of the three SCW HvCesA genes.
In terms of the “tight” co-regulation between the three endogenous HvCesA genes, there was a perturbation between eHvCesA4-eHvCesA8 (r2 = 0.2292) and eHvCesA7-eHvCesA8 (r2 = 0.0912) for plants with a ‘brittle node’ phenotype. Co-regulation of eHvCesA4-eHvCesA7 in the same plants remained tight (r2 = 0.8420). For all other plants with either stunted or normal phenotypes, the determination coefficient, r2, was in the range 0.53 to 0.86. This was quite different to the dwarfed SCW CesA transgenic lines and suggested that the brittle node phenotype may be a direct or indirect result of the perturbed co-regulation between eHvCesA8 and other eHvCesA genes. Furthermore, transcript profiles between normal and brittle node plants in T135S:HvCesA8 do not differ, which suggests that the aberrant phenotype is not associated with transcript abundance.
Crystalline cellulose content and stem strength
For T135S:HvCesA4 plants, there was no significant increase in cellulose content, as measured chemically, whether expressed as % cellulose per total cell wall (Figure 4A) or as mg cellulose per cm stem (data not shown). Normal-looking plants showed a flexural strength similar to the controls plants and most plants with dwarfism displayed a significant reduction in cellulose content and stem strength (Figures 4A and 4B). On average, cellulose per total cell wall decreased by 40% and the stem strength was also reduced to 20% of the average of control plants.
Figure 4.
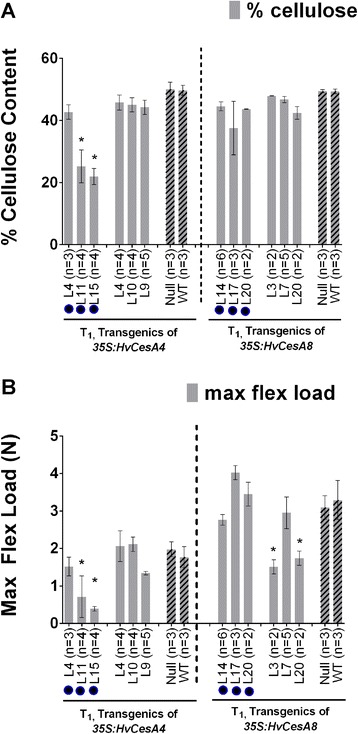
Cellulose content and stem strength data for T 1 SCW 35S: HvCesA4 and 35S: HvCesA8 plants. (A) Cellulose content was measured as percent cellulose (%). There were three independent lines with a dwarfed and leaf necrosis phenotype (black solid circle) and three lines with a normal phenotype. (B) maximum flexural load, N, was a measure of stem strength. There were three independent lines with ‘brittle node’ phenotype (black solid circle) and three normal-looking transgenic plants. Plants that were severely stunted died at a young stage so were not available for cellulose content analysis. Error bars are standard error of the mean of biological replicates (n). Significant differences were determined by one-way ANOVA followed by post hoc Dunnett’s multiple comparisons test.
Similarly, no significant increase in cellulose content for the 35S:HvCesA8 T1 plants was observed (Figure 4B), even where high levels of transgene transcript were detected, and there was no significant increase in plant stem strength. A 46-53% decrease in stem strength relative to controls was found in Line 3 and Line 20, which are likely due to reason unrelated to cellulose content.
T0 plants transformed with constructs PCW or SCW were also analysed and showed that there was no significant increase in either crystalline cellulose content or stem strength (data not shown).
Observation of crystalline cellulose using immunofluorescent- labelling in stem tissues of T2 plants
To examine potential changes in cellulose distribution as related to the chemically quantitated reduction shown in Figure 4, immunofluorescent labelling with the CBM3a protein was conducted, for both internode and node sections of dwarf T235S:HvCesA4 plants and brittle node T235S:HvCesA8 plants (Figure 5).
Figure 5.
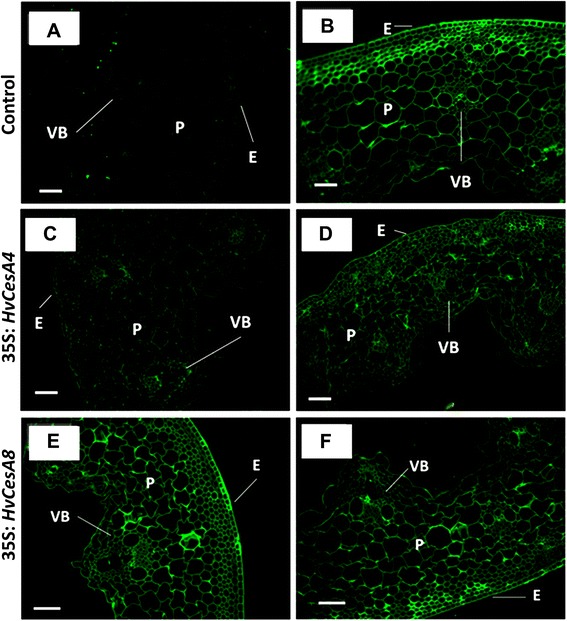
Immunofluorescent labelling of T 2 35S:HvCesA4 and T 2 35S:HvCesA8 internode cross-sections. (A) negative (same treatment as control and transgenic was applied but CBM3a was excluded), (B) control = wild type or nulls, (C) transgenic 35S:HvCesA4 plant from Line 11, (D) transgenic 35S:HvCesA4 plant from Line 15, (E) transgenic 35S:HvCesA8 plant from Line 14 and (F) transgenic plant from Line 20. Fluorescent images were taken at the same exposure and magnification for all samples. Scale bar is 100 μM. E = epidermis, VB = vascular bundle, PC = parenchyma cell.
Fluorescence intensity of labelling on both node and internode sections of T235S:HvCesA4 was reduced for all cell types (Figures 5C and 5D) compared with control sections (Figure 5B). In the case of the T235S:HvCesA8 plants (Figures 5E and 5 F), all cell types were labelled at a similar intensity to the control (Figure 5B). Similar reductions in intensity were detected in the node sections for T235S:HvCesA4 plants (Additional file 1: Figures S6 and S7).
Although the immunocytochemical images will normally give a semi-quantitative estimation of crystalline cellulose, the less intense fluorescence detected in the internode and node sections of T2 lines carrying the 35S:HvCesA4 construct was consistent with the reduced amounts of crystalline cellulose measured chemically (Figure 4).
Tissue architecture, cell wall thickness and lignin distribution in 35S: SCW HvCesA plants
Staining with toluidine blue showed that xylem vessels in internode cross-sections of dwarfed 35S:HvCesA4 T1 plants were partially collapsed and had irregular boundaries along the elliptical xylem vessels (Figure 6). Collapsed xylem vessels were also observed in leaves from dwarf plants (Additional file 1: Figure S8) but were not seen in 35S:HvCesA8 T1 brittle node plants or in control plants, where xylem vessels were round in shape (Figures 6A, 6B, 6E, 6F). For severely stunted 35S:HvCesA8 T1 plants, samples were collected and fixed shortly before the plant died. These plants appeared to comprise only the leaves arising from the crown of the base at the plant (Additional file 1: Figure S9). Secondary xylem (meta-xylem) did not develop, perhaps because the tissue was too young, but normal proto-xylem development was observed.
Figure 6.
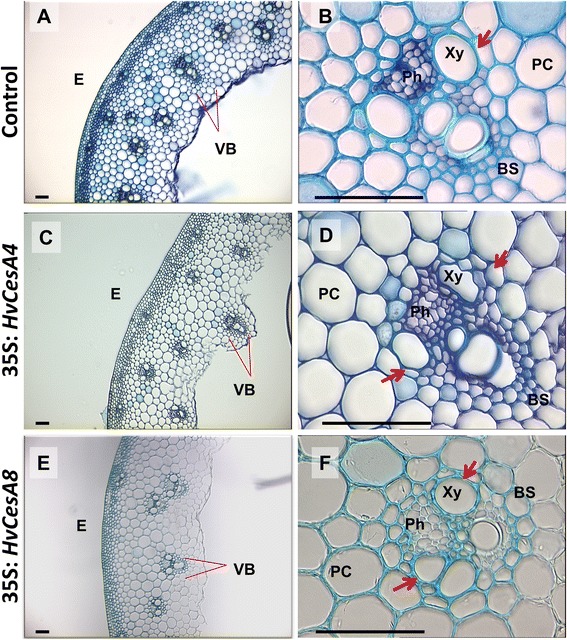
Light microscopy of cross-sections of T 1 35S:HvCesA4 and 35S:HvCesA8 stem internodes stained with Toluidine Blue. Equivalent internodes were sectioned using vibratome (~30-50 μM thick) from (A, B) wild-type or null, (C, D) dwarfed 35S:HvCesA4 transgenic T1 plants and (E, F) 35S:HvCesA8 transgenic T1 plants. Red arrows indicate xylem vessels and in D, they are collapsed and irregular in shape. Scale bars denote 100 μM. E = epidermis, VB = vascular bundle, Ph = phloem tissue, Xy = meta-xylem, BS = bundle sheath, PC = parenchyma cells.
Cell walls for collapsed xylem from T235S:HvCesA4 were examined for reductions in cell wall thickness, as were the sclerenchyma cells (Figures 7A – D). Consistent with the more severe morphology observed in 35S:HvCesA4 dwarf plants (i.e. collapsed xylem), their xylem cell walls were thinner overall, had irregular edges and were occasionally interrupted by apparent gaps in the middle lamella layer. In some cases, two walls detached at the middle lamella (Figure 7B). This was not seen in control plants. The SCW of sclerenchyma cells located under the epidermis of the stem also showed a reduced thickness for both T235S:HvCesA4 plants (Figure 7D). Cell walls in T235S:HvCesA4 appeared collapsed and cell wall thickenings were located mainly at cell corners.
Figure 7.
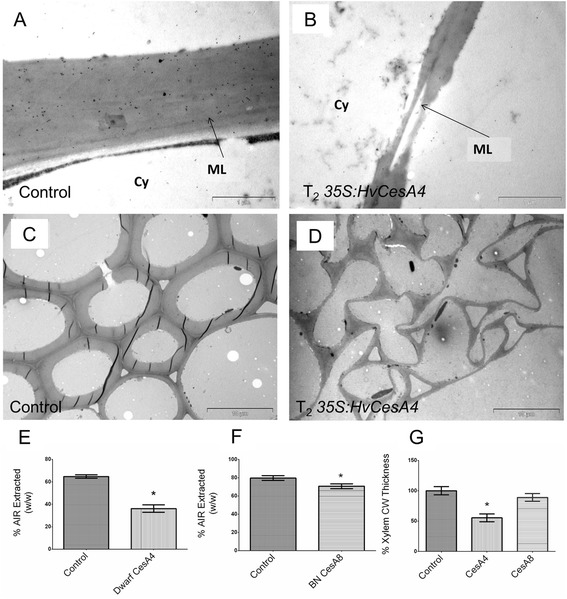
TEM and measurement of SCW thickening for control and T 2 transgenic plants. (A) Xylem cell wall of WT, (B) Xylem cell wall of T2 dwarf 35S:HvCesA4, (C) Sclerenchyma cell wall of WT and (D) Sclerenchyma cell wall of T2 dwarf 35S:HvCesA4. Scale bar is 1 μM for (A, B) and 10 μM for (C, D). Cy = Cytoplasm of bundle sheath cell, ML = middle lamella. (E, F) Percent AIR extracted (w/w) from 35S:HvCesA4, 35S:HvCesA8 and control from stem tissues. (G) % reduction of xylem cell wall thickness as measured using ImageJ.
Measurements for xylem cell wall thickness were taken from ten images of xylem vessels imaged from two independent lines and a 45% reduction in xylem cell wall thickness was found in plants carrying 35S:HvCesA4 (Figure 7G). This decrease in cell wall thickness was further supported by a decrease in percentage of total cell wall material (AIR) extracted from stem tissues, although we acknowledge that the yield of AIR material will be only semi-quantitative in nature. It was found that dwarf 35S:HvCesA4 and brittle node 35S:HvCesA8 plants had 44% and 10% reductions in total AIR extracted, respectively (Figures 7E, 7F).
Aohara et al. [24] attributed a rice “brittle node” phenotype to a drastic reduction of lignified tissues in the node. Nodes from three different T235S:HvCesA4 and 35S:HvCesA8 plants exhibiting the ‘brittle node’ phenotype were therefore sectioned and stained with phloroglucinol-HCl but no significant changes in lignin content were observed (Additional file 1: Figure S10).
Discussion
To investigate whether stem strength in barley and hence resistance to lodging might be improved through increasing cellulose levels in cell walls, barley was transformed with individual PCW (HvCesA1, HvCesA2, HvCesA6) and SCW (HvCesA4 and HvCesA8) cellulose synthase cDNAs from barley (Burton et al., [15]), driven by the constitutive CaMV 35S promoter. We have used the CaMV 35S promoter successfully to over-express transgenes in barley and other groups have used this promoter to successfully over-express transgenes in rice [25-27], although we acknowledge that alternative promoters such as maize ubiquitin [28] and rice actin [29] have been shown to be generally more active in monocots.
Visual phenotypes
The HvCesA transgenic plants were examined for a visual phenotype, transcript levels of both transgenes and endogenous genes, stem strength, stem morphology, cell wall ultrastructure, cellulose content and crystalline cellulose distribution in the cell wall. More than 90% of T0 transgenic plants carrying PCW 35S:HvCesA constructs exhibited no drastic phenotypic defects but aberrant phenotypes were observed in approximately 25% of the SCW 35:HvCesA plants and these phenotypes persisted into the T1 and T2 generations (Figure 2). From the transcript analyses of T0 PCW 35S:HvCesA plants, transgene expression levels for all individuals were very low and in some transgenic populations, the endogenous genes were co-suppressed (Additional file 1: Figures S1-S5). For SCW 35S:HvCesA plants, transgene transcript levels higher than the endogenous transcripts in control plant were found (Figure 3) but this did not result in any significant increase in cellulose content above control levels (Figure 4). Indeed, we were unable to increase the total cellulose content in any of the transgenic lines and in some lines it decreased significantly (Figure 4).
Reductions in crystalline cellulose
The present work showed that the crystalline cellulose content of 35S:HvCesA4 dwarf plants, as determined by the Updegraff [30] method, was lower than control plants (Figure 4). The reduction in crystalline cellulose was confirmed in both nodes and internodal regions of the stem by immunofluorescence (Figure 5). A reduction in crystalline cellulose may not be the sole contributor to the defect in xylem integrity; a reduction in lignin might also be a contributing factor. Phloroglucinol-HCl staining of xylem cells indicated that although lignin was present, no large differences could be detected between the control and transgenic plants. To quantitate more subtle reductions in lignin content, Klasson lignin assays [31] could be used, but these assays were not applied in the present study.
Common perturbations in cell morphology
In dwarfed SCW 35S:HvCesA4 barley lines, cell morphology was perturbed and xylem vessels had both collapsed and showed a reduction in cell wall thickness (Figures 6 and 7). A similar phenotype has been described in Arabidopsis irx mutants [32]. When Joshi and collaborators [33] introduced another copy of the SCW Populus tremuloides L cellulose synthase PtdCesA8 gene, which is the putative orthologue of barley HvCesA7, driven by the CaMV 35S promoter into transgenic poplar plants, severe silencing of both the endogenous and transgene CesAs, together with a dramatically reduced cellulose content, dwarfism and a collapsed xylem phenotype, were observed. However, Joshi et al. [33] did not report a reduction in wall thickness. In contrast, reductions in cell wall thickness were detected here in T235S:HvCesA4 plants that exhibited a dwarf phenotype, where the reduction in xylem cell wall thickness was accompanied by an apparent reduction in total extractable cell wall material (Figure 7).
The striking resemblance between phenotypes (collapsed xylem, dwarfism, early leaf senescence) for the 35S:HvCesA4 construct in barley and the cellulose deficient 35S:CesA transgenic poplar further strengthen the suggestion that, despite the wide phylogenetic distance between a woody tree and a grass, the regulation of SCW CesAs may be conserved. However, it is still unclear if the phenotypic changes observed in the barley transgenic lines are directly attributable to silencing the corresponding SCW HvCesA4 gene or to pleiotropic effects, because mutations other than those in CesA genes invoke similar morphological defects. Examples are mutations in genes involved in lignin biosynthesis [34,35], xylan biosynthesis [36], a mutated endoglucanase [37] and pectin biosynthesis via over-expression of QUA2 [38], which all resulted in collapsed xylem vessels.
Tight regulation and different effects are observed for individual HvCesA genes
Our results demonstrated that perturbing HvCesA gene expression in the some transgenic lines not only caused extreme phenotypes but also resulted in the silencing of endogenous HvCesA genes and, in many cases, in reduced crystalline cellulose contents. It appears likely therefore that barley, and probably other plants, have evolved tight regulatory mechanisms to maintain cellulose levels within a relatively narrow range. Studies in transgenic petunia and other plants indicate that sense co-suppression can be related to promoter strength [39]. However, in the present study, some transgenic lines showed similar or higher endogenous HvCesA transcript levels compared with the control plants, but displayed the same phenotypic features as the lines in which transcript levels were lower.
The 35S:HvCesA4 construct caused more severe developmental defects than the 35S:HvCesA8 construct. The T2 transgenic plants of both constructs were brittle at the nodes but 35S:HvCesA4 plants were also dwarfed and had collapsed xylem vessels. The differences in the severity of the transgenic phenotypes between 35S:HvCesA4 and 35S:HvCesA8 suggest that the protein products of these two secondary cell wall HvCesA genes have different or unequal roles in cellulose synthesis. It has been shown that in the fs2 brittle stem mutant of barley, in which transcription of the HvCesA4 gene is compromised by the presence of a retrotransposon in the first intron of the gene, cellulose crystallinity is reduced [40]. However, the tight co-regulation between the two groups of three endogenous HvCesA genes was not perturbed in the fs2 brittle stem mutant of barley. In contrast, the tight co-expression of these genes was not always retained in transgenic lines generated in the present study, in which plants with a ‘brittle node’ phenotype showed much reduced co-efficients of determination between eHvCesA4-eHvCesA8 and eHvCesA7-eHvCesA8. Co-regulation of eHvCesA4-eHvCesA7 in the same plants remained tight (r2 = 0.8420). For all other plants with either stunted or normal phenotypes, r2 was in the range 0.53 to 0.86. In contrast to the situation in the fs2 brittle stem mutant [40], the ‘brittle node’ phenotype observed here may be a direct or indirect result of a breakdown of the co-regulation of the eHvCesA8 gene and genes encoding its putative partners in the cellulose synthase complex.
There is some evidence of redundancy and dual functionality in the roles of CesA proteins in Arabidopsis, where the PCW AtCesA2 and AtCesA5 proteins are partially redundant to AtCesA6, one of the three CesA protein subunits in the PCW cellulose synthase complex [18,41,42]. Moreover, the Arabidopsis SCW AtCesA7, which is the putative orthologue of the barley SCW HvCesA8, was found to partially rescue defects in the PCW mutant cesa3 and, conversely, PCW AtCesA1 was able to partly rescue defects in the SCW mutant cesa8ko [43]. This suggests that there is flexibility between CesA protein function depending on the tissue or environmental conditions [44]. It might also be argued that the constitutive expression of the 35S promoter leads to a negative dominant phenotype by disturbing the endogenous gene expression in the secondary wall forming cells. Mis-assembly of the functional enzyme complex may ensue because of disturbed stoichiometry of the various subunits. The transcript profiles from this study also showed that stoichiometry is maintained, pointing to the existence of a homeostatic mechanism. The HvCesA8 gene showed strong overexpression in most lines and was associated with the downregulation of endogenous genes in lines with dwarf phenotypes. The brittle phenotype was correlated with high transgene expression in the absence of strongly reduced transcript levels of endogenous CesA genes. This suggests that the perturbation of the stoichiometry of individual CesA proteins in the cellulose synthase complex (CSC) may be responsible for the brittle phenotype.
Brittle stems arise by different genetic lesions
In the fs2 barley mutant, a brittle stem phenotype arose due to the insertion of a retroelement in the first intron of the HvCesA4 gene. This fs2 mutant had reduced crystalline cellulose and increased non-crystalline cellulose compared to control plants [40]. This showed that HvCesA4 is essential for the integrity of the cell wall that would otherwise lead to brittleness of the stem. In the present study, brittleness caused by SCW 35S:HvCesA4 was restricted to the node region of the plant stem, unlike the brittleness found in the fs2 mutant [40]. The less severe phenotype may arise due to the presence of low level of functional CesA4 protein in the silenced transgenic lines described here, whereas in the fs2 mutant, there may have been no functional CesA4 protein present. As to why the phenotype was restricted to the nodal region, perhaps these joints are the weakest point of the stem and any cell wall defects would be more obvious in this area.
The “brittle node” phenotype exhibited by both 35S:HvCesA4 and 35S:HvCesA8 plants also resembled the phenotype previously described in the rice bc5 brittle culm mutant [24]. The exact gene affected by this mutation was not identified but SCW OsCesA transcripts were suppressed and it was found that walls of sclerenchyma cells in the leaf sheath bundle around the stem node were thinner [24]. Although suppression of all SCW eHvCesA genes was also found in the “brittle node” plants in the present study, gene suppression alone cannot explain the “brittle node” phenotype observed in barley because other transgenic plants with a normal phenotype displayed similar suppression of transcripts. The “brittle node” phenotype may be caused by the disrupted coordination between HvCesA4 and HvCesA8 proteins as a result of altered transcript levels, although other factors such as the formation of the rosette complex, cellulose assembly and interactions with other players in cell wall assembly are also likely to affect the final phenotype of a plant.
Our inability to increase cellulose content in the transgenic barley lines may also be attributable to a requirement for all three HvCesA genes in a complex to be over-expressed simultaneously. Thus, concurrent up-regulation may be needed to produce a successful increase in cellulose. Although attempts to simultaneously up-regulate the three co-ordinately expressed PCW and SCW HvCesA genes were not within the scope of this paper, such a goal using the current lines is possible in the future.
Conclusions
In summary, the results presented here indicate that potential challenges could be encountered in attempts to engineer cellulose levels in planta by manipulating either PCW or SCW HvCesA genes using the CaMV 35S promoter. However, the observed pleiotropic phenotypes and transcript silencing arising from our systematic introduction of individual HvCesA genes into transgenic barley provided us with an opportunity to deduce the roles of individual HvCesA genes. Our results demonstrated unequal roles within SCW genes and between PCW and SCW genes to maintain the structural integrity of cell walls and on the overall ability of the plants to stay upright. The HvCesA4 gene showed the most negative effects on plant growth in barley. Similar developmental defects observed by Joshi and colleagues [33] for a tree species indicate tight regulation of cellulose biosynthetic genes occurs across the plant kingdom and further work is needed to unravel the complexity of this process.
Methods
In Silico mapping of CesA genes in grasses
Barley, rice and sorghum CesA gene positions were estimated from the barley scaffold genome sequence [22,45] and Ensembl Plants (http://plants.ensembl.org/info/website/ftp/index.html) [46] and mapped using Strudel, a stand-alone Java desktop application that allows the simultaneous multi-way comparison of several genomes (http://bioinf.scri.ac.uk/strudel/; [47]).
Vector construction
Three PCW and two SCW CesA genes were individually cloned into the pMDC32 vector [47]. The forward and reverse primer pairs used to clone each full length gene are given in Table S1. Full length HvCesA1, HvCesA2, HvCesA4, HvCesA6 and HvCesA8 cDNAs were generated as described in Burton and co-authors [15] and were cloned into the pCR8®/GW/TOPO TA vector (Life Technologies, Australia). Clones from each construct were digested with restriction enzymes to select those with a sense orientation and were subsequently sequenced on an ABI 3700 (Applied Biosystems Inc., Australia) at the Australian Genome Research Facility (Adelaide, Australia) to verify the identity of genes and the precision of constructs. Each HvCesA cDNA was transferred (Life Technologies, Australia) into a Gateway-enabled constitutive expression vector, pMDC32 [48], carrying dual 35S promoters and a NOS terminator that flank the inserted CesA cDNA at the 5′ and 3′ ends of the gene, respectively.
Barley transformation and plant material
A total of five constructs were individually transformed into Agrobacterium tumefaciens strain AGL-1, using the freeze-thaw method [49]. The procedures used to grow barley donor plants (Hordeum vulgare cv. Golden Promise) and prepare immature scutella for transformation were previously described in Burton et al. [25]. The scutella were cultured on callus induction medium in the dark at room temperature for a day prior to transformation. The constructs were transformed into barley using the protocol developed by Tingay et al. [50] and modified by Matthews et al. [51].
The T0 and T1 transgenic plants were grown under standard glasshouse conditions as described in Burton and colleagues [15] while T2 plants were grown in The Plant Accelerator (Australian Plant Phenomics Facility) under the same conditions. Transgenic plants of T135S:HvCesA4 and 35S:HvCesA8 were grown at different times of the year, thus direct comparison of transcript and cellulose data are made only between transgenic and control plants. Transgenic plants and their comparable control plants were grown in the same growth chamber and the control plants were part of a random design.
Fertile T0 transgenic plants, which exhibited either normal or aberrant phenotypes, and further contained a low locus number of the transgene (one to two), were selected to grow on to the T1 and T2 generations. The presence of the transgene in T1 and T2 plants was confirmed using a Phire® Plant Direct PCR Kit (Finnzymes, Vantaa, Finland) and melt curve PCR, respectively. In addition, random T1 transgenic plants were selected for Southern hybridisation to verify the presence of the transgene (data not shown).
RNA isolation, cDNA synthesis and transcript analysis
Total RNA was extracted from leaves of 6-week-old barley (cv Golden Promise) using a commercially prepared guanidine reagent, TRIzol (Life Technologies, Australia) following the manufacturer’s instructions. All RNA samples obtained were treated with the TurboDNA-free DNAse kit (Ambion) to remove genomic DNA contamination. The procedure used for cDNA synthesis was that described by Burton et al. [15].
Real time quantitative PCR (QPCR) amplification of the 3′ untranslated region (UTR) of HvCesA genes was conducted with gene-specific primers as described in Burton et al.[15]. Primers used to detect transgenes are listed in Additional file 1: Table S2. It was possible to distinguish between endogenous and transgene HvCesA DNA because the 3′UTR of the transgene was shorter and lacked the endogenous reverse primer binding site. The reverse primer for the transgene was designed to hybridize to the NOS terminator, which is not present in a wild-type gene (Additional file 1: Figure S6). The amplified QPCR products were subsequently sequenced to verify the identity of the gene fragment. QPCR was carried out as in Burton et al. [52] and the cDNA population used for the initial amplification to determine the acquisition temperatures of QPCR products (Additional file 1: Table S2) was a mixed population of cDNAs synthesized from a cocktail of RNAs extracted from different transgenic plants.
The same control cDNAs were used for QPCR analysis of all five sets of T0 transgenic plants. Control cDNAs for T1 SCW 35S:HvCesA plants were null and wild type plants were sown together with the transgenic plants. As a control for non-specific binding, Q-PCR was conducted to detect transgene transcript levels in wild type cDNAs but only background levels were observed.
Microscopy
Stem sections of at least three T1 plants from each line of plants transformed with 35S:HvCesA4 or 35S:HvCesA8 were fixed in freshly prepared Farmer’s fixative (ethanol:acetic acid = 3:1) for 24 hours and transferred to 70% ethanol at 4°C. For all plants, the second stem internode below the flag leaf was sectioned, except for stunted plants that died before proper stem development.
For general morphological studies, fixed internode tissues were briefly rinsed with water and sectioned with a Leica VT1200 vibratome (30–50 μm thickness, 0.4 mm/s velocity, 0.4 mm amplitude). Sections were stained with 0.05% toluidine blue in benzoate buffer pH 4.4, for 2 min and rinsed with water seven times. To obtain uniform sections for soft stems (plants that died young) and ensure comparability, all stem pieces from 35S:HvCesA8 lines were further dehydrated through a series of ethanol concentrations, embedded in paraffin wax and sectioned on a RM 2155 Microtome (Leica) to a thickness of 7 μm. After dewaxing and rehydration, the sections were stained with 0.05% toluidine blue.
To examine lignin, Farmer’s fixed sections of nodes and internodes were stained with 2% (w/v) phloroglucinol (Sigma) in 95% ethanol, followed by a water rinse before treatment with hydrochloric acid. Experiments were carried out as described in Liljegren [53]. All images were captured on Leica AS LMD Laser Dissection Microscope.
Stem strength measurement
Five stems per plant were tagged during plant development. Stems used for measurements were from the five tallest tillers from the plant. The primary tiller was not used because it is usually the strongest and therefore is different in strength to the secondary tillers. The barley transgenic T0 and T1 lines were grown in a glasshouse until maturity and air dried stem samples (7-8% moisture content) were used for stem strength measurements. An Instron 5543 materials testing instrument was used to test for three-point flexural strength at a span distance of 40 mm and an anvil rate of 60 mm min−1 and data were analysed using Bluehills 2 material testing software. Measurements for flexure load at break (N) were taken at the third internode below the flag leaf. The flexural load (N) required to bend the midpoint of each internode was recorded. The average flexure load for three out of the five internodes with the most similar diameters was calculated. Other measurements such as stem diameter and internode length were also recorded. All transgenic, null and wild type plants were tested.
Cell wall preparations and crystalline cellulose assay
Freeze dried stems from segments of three internodes directly below the flag leaf were ground to a powder using a Spex 2000 GenoGrinder [2–5 min at 500 (1500strokes/min)] and alcohol-insoluble residues (AIR) were extracted in accordance with Zhang and co-authors [54]. A modified Updegraff (acetic/nitric acid cellulose assay) [30] method, described in Burton and co-authors [40], was performed to determine crystalline cellulose content (% stem or cell wall weight) and mg of cellulose per cm stem. Cellulose assay for all T1 transgenics and controls were determined for AIR (alcohol insoluble residues) material. All available T1 plants were tested with at least three independent lines for each construct. Cellulose value for each plant was calculated from measurements obtained from three separate tillers per plant. For each cellulose assay, coefficient of variance for technical replicates was less than 15%.
Immunofluorescent labelling with CBM3a by light microscopy
Barley stem sections (1 cm internode and node) from T2 35S:HvCesA4 and 35S:HvCesA8 plants were fixed overnight at 4°C in 4% sucrose, 4% paraformaldehyde and 0.25% glutaraldehyde. Samples were washed with phosphate buffered saline (PBS), pH 7.4 and dehydrated in an increasing series of ethanol concentrations (70%, 80%, 90% 95%, 100%) and finally embedded in LR white resin (ProSciTech, Australia). Sections of 1 μm thickness were cut on a Leica Ultracut R microtome using a diamond knife and affixed onto Poly-L-Lysine glass microscopy slides (Thermo Scientific, Australia). A three-stage immunolabelling method was conducted as described in McCartney [55] with the following modifications optimised for barley stem sections. Specimens were incubated with 6.25 μg/mL CBM3a (Plant Probes, Leeds, UK) [56] in 1% bovine serum albumin/phosphate-buffered saline (BSA/PBS), pH7.4 for 1 h, followed by a 1:100 dilution of the mouse anti-Histidine (His) monoclonal antibody (Sigma-Aldrich, Australia) for 1.5 h and a 1:100 dilution of Alexa Fluor 488 goat anti-mouse IgG (Life Technologies, Australia) for 1 h in the dark. Samples were washed with PBS, mounted in 90% glycerol and viewed on a Leica AS LMD Laser Dissection Microscope with a DFC 480 camera, using fluorescence filter I3 (excitation filter 450–490 nm BP, barrier filter 515 nm LP). For each construct at least three plants from two independently transformed lines were imaged.
In control experiments, the specimen was incubated with a combination of mouse anti-His monoclonal antibody and Alexa Fluor 488 goat anti-mouse IgG, while the CBM3a protein was omitted. Results from the immunofluorescent labelling are comparable because cross-sections from control and transgenic plants were treated on the same slide under the same conditions. At least two plants from two independent lines were examined and all images were captured at the same exposure.
Transmission Electron Microscope (TEM)
Two plants, each from a single independent line, were examined for each transgenic construct. Null and wild type plants were examined as controls. Sample preparation was as described for immunofluorescent labelling except sections of 80 nm were cut and collected on coated nickel grids (200 mesh parallel). Grids were stained with 2% (w/v) aqueous uranyl acetate (Sigma-Aldrich, Australia) before images were taken using a Philips (Eindhoven, The Netherlands) BioTwin transmission electron microscope and a Gatan multiscan digital camera. The mean of cell wall thickness for each construct was estimated from 10 images representing xylem cells from two different independent lines using ImageJ [57].
Statistical analysis
All statistical analyses such as ANOVA and correlation analysis were performed using GraphPad Prism (GraphPad Software, Inc., California).
Availability of supporting data
Sequence data from this article can be found in the GenBank data libraries under accession numbers [AY483150: HvCesA1, AY483152: HvCesA2: HM222644: HvCesA4, AY483155: HvCesA6 and KM45970: HvCesA8]. All the supporting data are included as additional files.
Acknowledgements
We are grateful to Dr Jacinda Rethus for vector construction, Adelaide Microscopy (North Terrace) for the use of TEM and Dr Ursula Langridge, Ms Kylie Neumann and Mr Kuok Cheang Yap for additional help in maintaining and harvesting the plants. We are also thankful to Ms Jillian Taylor for providing training on the Instron machine.
Abbreviations
- CesA
Cellulose synthase genes
- PCW
Primary cell wall
- SCW
Secondary cell wall
- HvCesA
Barley cellulose synthase
- FTIR
Fourier transform infrared resonance
- PCA
Principle component analysis
- CBM3a
Crystalline cellulose-binding module (recombinant CBM protein)
- QPCR
Real time quantitative PCR
- AIR
Alcohol insoluble residues
- PBS
Phosphate buffered saline
Additional file
Ratios of transgene/endogenous control levels and the percentage reductions of endogenous transcript levels (average value). Table S2. Primers used for cloning HvCesA full length genes with 5′ and 3′ends. Table S3. QPCR primers for transgene. Figure S1. Box plot graphs showing transcript profiles for all 35S:HvCesA1 plants. Figure S2. Box plot graphs showing transcript profiles for all 35S:HvCesA2 plants. Figure S3. Box plot graphs showing transcript profiles for all 35S:HvCesA6 plants. Figure S4. Box plot graphs showing transcript profiles for all 35S:HvCesA4 plants. Figure S5. Box plot graphs showing transcript profiles for all 35S:HvCesA8 plants. Figure S6. Immunofluorescent labelling of T2 35S:HvCesA4 node cross-sections. Figure S7. Immunofluorescent labelling of T2 35S:HvCesA8 node cross-sections. Figure S8. Partially collapsed xylem vessels found in vibratome sections. Figure S9. Light microscopy of cross-sections of 35S:HvCesA8 stem internodes stained with Toluidine Blue. Figure S10. Bright field microscopy of phloroglucinol-HCl stained node and internode cross-sections. Figure S11. Schematic representation of the primer binding sites for endogenous and transgene HvCesAs.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HT, GF and RB conceived and designed the experiments, HT, NS, RS, MH and GM performed the experiments, HT, NS, GF, KD and RB analysed and interpreted the data, HT, GF and RB wrote and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Hwei-Ting Tan, Email: hwei.tan@adelaide.edu.au.
Neil J Shirley, Email: neil.shirley@adelaide.edu.au.
Rohan R Singh, Email: rohan.singh@adelaide.edu.au.
Marilyn Henderson, Email: marilyn.henderson@adelaide.edu.au.
Kanwarpal S Dhugga, Email: dhuggaks@gmail.com.
Gwenda M Mayo, Email: gwenda.mayo@adelaide.edu.au.
Geoffrey B Fincher, Email: Geoff.fincher@adelaide.edu.au.
Rachel A Burton, Email: rachel.burton@adelaide.edu.au.
References
- 1.Berry PM, Sterling M, Mooney SJ. Development of a model of lodging for barley. J Agron Crop Sci. 2006;192(2):151–8. doi: 10.1111/j.1439-037X.2006.00194.x. [DOI] [Google Scholar]
- 2.Berry PM, Sterling M, Spink JH, Baker CJ, Sylvester-Bradley R, Mooney SJ, Tams AR, Ennos AR. Understanding and reducing lodging in cereals. Adv Agronomy. 2004;84:217–71. doi: 10.1016/S0065-2113(04)84005-7. [DOI] [Google Scholar]
- 3.Rüggeberg M, Speck T, Burgert I. Structure–function relationships of different vascular bundle types in the stem of the Mexican fanpalm (Washingtonia robusta) New Phytol. 2009;182(2):443–50. doi: 10.1111/j.1469-8137.2008.02759.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Ren H, Zhang B, Fei B, Burgert I. Cell wall structure and formation of maturing fibres of moso bamboo (Phyllostachys pubescens) increase buckling resistance. J R Soc Interface. 2012;9(70):988–96. doi: 10.1098/rsif.2011.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rüggeberg M, Burgert I, Speck T. Structural and mechanical design of tissue interfaces in the giant reed Arundo donax. J R Soc Interface. 2010;7(44):499–506. doi: 10.1098/rsif.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kokubo A, Kuraishi S, Sakurai N. Culm strength of barley. Plant Physiol. 1989;91(3):876–82. doi: 10.1104/pp.91.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma QH, Xu Y, Lin ZB, He P. Cloning of cDNA encoding COMT from wheat which is differentially expressed in lodging-sensitive and resistant cultivars. J Exp Bot. 2002;53(378):2281–2. doi: 10.1093/jxb/erf102. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003;133(1):73–83. doi: 10.1104/pp.103.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, et al. Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta. 2006;224(5):1174–84. doi: 10.1007/s00425-006-0299-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Zhu J, Huang RZ, Yang YS. Investigation of cell wall composition related to stem lodging resistance in wheat (Triticum aestivum L.) by FTIR spectroscopy. Plant Signal Behav. 2012;7(7):1–8. doi: 10.4161/psb.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Appenzeller L, Doblin M, Barreiro R, Wang H, Niu X, Kollipara K, et al. Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose. 2004;11(3):287–99. doi: 10.1023/B:CELL.0000046417.84715.27. [DOI] [Google Scholar]
- 12.Brown RM., Jr Cellulose structure and biosynthesis: what is in store for the 21st century? J Polymer Sci Part A Polymer Chem. 2004;42(3):487–95. doi: 10.1002/pola.10877. [DOI] [Google Scholar]
- 13.Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50(1):245–76. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 14.Guerriero G, Fugelstad J, Bulone V. What do we really know about cellulose biosynthesis in higher plants? J Integr Plant Biol. 2010;52(2):161–75. doi: 10.1111/j.1744-7909.2010.00935.x. [DOI] [PubMed] [Google Scholar]
- 15.Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 2004;134(1):224–36. doi: 10.1104/pp.103.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12(12):2529–40. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci U S A. 2003;100(3):1450–5. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci. 2007;104(39):15572–7. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22(1):53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 20.Perrin RM. Cellulose: how many cellulose synthases to make a plant? Curr Biol. 2001;11(6):R213–6. doi: 10.1016/S0960-9822(01)00108-7. [DOI] [PubMed] [Google Scholar]
- 21.Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell Online. 2011;23(4):1249–63. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium IBGS. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–6. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 23.Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Res. 1974;14(6):415–21. doi: 10.1111/j.1365-3180.1974.tb01084.x. [DOI] [Google Scholar]
- 24.Aohara T, Kotake T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S. Rice brittle culm 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant Cell Physiol. 2009;50(11):1886–97. doi: 10.1093/pcp/pcp133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton RA, Collins HM, Kibble NAJ, Smith JA, Shirley NJ, Jobling SA, et al. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol J. 2011;9(2):117–35. doi: 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- 26.Katsuhara M, Koshio K, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K. Over-expression of a barley aquaporin increased the shoot/root ratio and raised salt sensitivity in transgenic rice plants. Plant Cell Physiol. 2003;44(12):1378–83. doi: 10.1093/pcp/pcg167. [DOI] [PubMed] [Google Scholar]
- 27.Battraw M, Hall T. Histochemical analysis of CaMV 35S promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Mol Biol. 1990;15(4):527–38. doi: 10.1007/BF00017828. [DOI] [PubMed] [Google Scholar]
- 28.Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5(3):213–8. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 29.McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell Online. 1990;2(2):163–71. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Updegraff DM. Semimicro determination of cellulose inbiological materials. Anal Biochem. 1969;32(3):420–4. doi: 10.1016/S0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 31.Theander O, Westerlund EA. Studies on dietary fiber. 3. Improved procedures for analysis of dietary fiber. J Agric Food Chem. 1986;34(2):330–6. doi: 10.1021/jf00068a045. [DOI] [Google Scholar]
- 32.Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell Online. 1997;9(5):689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi CP, Thammannagowda S, Fujino T, Gou J-Q, Avci U, Haigler CH, et al. Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen. Mol Plant. 2011;4(2):331–45. doi: 10.1093/mp/ssq081. [DOI] [PubMed] [Google Scholar]
- 34.Piquemal J, Lapierre C, Myton K, O’connell A, Schuch W, Grima-pettenati J, et al. Down-regulation of Cinnamoyl-CoA Reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 1998;13(1):71–83. doi: 10.1046/j.1365-313X.1998.00014.x. [DOI] [Google Scholar]
- 35.Jones L, Ennos AR, Turner SR. Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 2001;26(2):205–16. doi: 10.1046/j.1365-313x.2001.01021.x. [DOI] [PubMed] [Google Scholar]
- 36.Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, et al. The arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell Online. 2007;19(1):237–55. doi: 10.1105/tpc.106.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szyjanowicz PMJ, McKinnon I, Taylor NG, Gardiner J, Jarvis MC, Turner SR. The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 2004;37(5):730–40. doi: 10.1111/j.1365-313X.2003.02000.x. [DOI] [PubMed] [Google Scholar]
- 38.Fuentes S, Pires N, Østergaard L. A clade in the QUASIMODO2 family evolved with vascular plants and supports a role for cell wall composition in adaptation to environmental changes. Plant Mol Biol. 2010;73(6):605–15. doi: 10.1007/s11103-010-9640-5. [DOI] [PubMed] [Google Scholar]
- 39.Que Q, Wang HY, English JJ, Jorgensen RA. The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell Online. 1997;9(8):1357–68. doi: 10.1105/tpc.9.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burton RA, Ma G, Baumann U, Harvey AJ, Shirley NJ, Taylor J, et al. A customized gene expression microarray reveals that the brittle stem phenotype fs2 of barley is attributable to a retroelement in the HvCesA4 cellulose synthase gene. Plant Physiol. 2010;153(4):1716–28. doi: 10.1104/pp.110.158329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci U S A. 2001;98(18):10079–84. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(39):15566–71. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carroll A, Mansoori N, Li S, Lei L, Vernhettes S, Visser RGF, et al. Complexes with mixed primary and secondary cellulose synthases are functional in Arabidopsis thaliana plants. Plant Physiol. 2012;160(2):726–37. doi: 10.1104/pp.112.199208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mutwil M, Debolt S, Persson S. Cellulose synthesis: a complex complex. Curr Opin Plant Biol. 2008;11(3):252–7. doi: 10.1016/j.pbi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, et al. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ) Plant J. 2013;76(4):718–27. doi: 10.1111/tpj.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kersey PJ, Allen JE, Christensen M, Davis P, Falin LJ, Grabmueller C, et al. Ensembl genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42(D1):D546–52. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayer M, Milne I, Stephen G, Shaw P, Cardle L, Wright F, et al. Comparative visualization of genetic and physical maps with Strudel. Bioinformatics. 2011;27(9):1307–8. doi: 10.1093/bioinformatics/btr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–9. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18(4):675–89. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 50.Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thornton S, et al. Agrobacterium tumefaciens‐mediated barley transformation. Plant J. 1997;11(6):1369–76. doi: 10.1046/j.1365-313X.1997.11061369.x. [DOI] [Google Scholar]
- 51.Matthews PR, Wang M-B, Waterhouse PM, Thornton S, Fieg SJ, Gubler F, et al. Marker gene elimination from transgenic barley, using co-transformation with adjacenttwin T-DNAs’ on a standard Agrobacterium transformation vector. Mol Breeding. 2001;7(3):195–202. doi: 10.1023/A:1011333321893. [DOI] [Google Scholar]
- 52.Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, et al. The genetics and transcriptional profiles of the cellulose synthase-Like HvCslF gene family in Barley. Plant Physiol. 2008;146(4):1821–33. doi: 10.1104/pp.107.114694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liljegren S. Phloroglucinol stain for lignin. Cold Spring Harbor Protocols 2010; doi:10.1101/pdb.prot4954. [DOI] [PubMed]
- 54.Zhang Q, Pettolino FA, Dhugga KS, Rafalski JA, Tingey S, Taylor J, et al. Cell wall modifications in maize pulvini in response to gravitational stress. Plant Physiol. 2011;156(4):2155–71. doi: 10.1104/pp.111.179606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCartney L, Gilbert HJ, Bolam DN, Boraston AB, Knox JP. Glycoside hydrolase carbohydrate-binding modules as molecular probes for the analysis of plant cell wall polymers. Anal Biochem. 2004;326:49–54. doi: 10.1016/j.ab.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 56.Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, et al. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281(39):29321–9. doi: 10.1074/jbc.M605903200. [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Meth. 2012;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


