Figure 1.
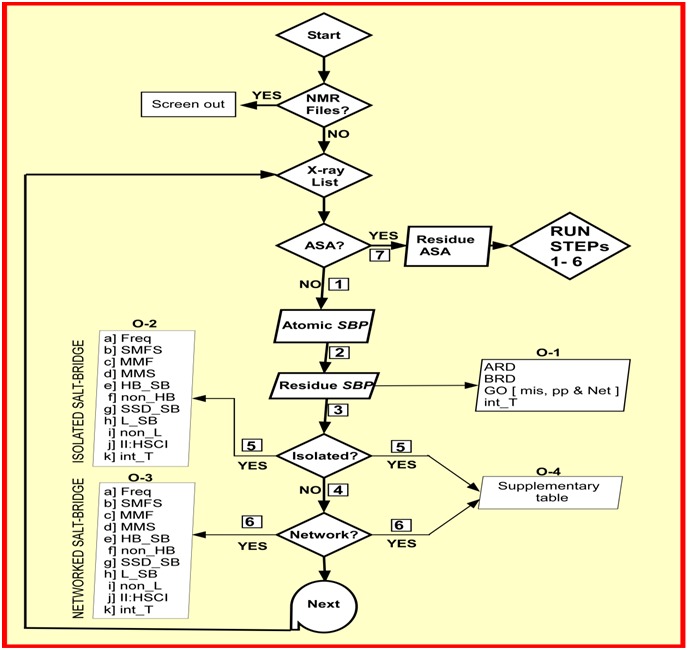
Flowchart of the functioning of the program SBION2. Upon start of the program it screens out NMR files if they are there in the working directory followed by making a list of X-ray structure files. The program verify for accessibility calculation program (ASA) in specified directory. If absent (i.e. NO), the program obtains atomic salt bridge pairs (step-1) and then converts them in residue specific salt bridge pairs (step-2). At this level four outputs are redirected (O-1). Output details of isolated (O-2) and networked salt-bridges (O-3) are separately reported (STEPs 3 to 6). A supplementary table (O-4) is prepared which includes both isolated and networked salt-bridge information (STEPs 5 and 6). If ASA (STEP 7) is YES, SBION2 performs step 1 through 6 that include core and surface location information of salt bridges along with earlier ones. Freq: Frequency; SMFS: Single bond multiplicity frequency and sum; MMF: Multiple bond multiplicity frequency; MMS: Multiple bond multiplicity sum; HB_SB: Hydrogen bonded salt-bridge frequency; non_HB: non-Hydrogen bonded salt-bridge frequency; SSD_SB: secondary structure (helix, sheet and coil) distribution of salt-bridge; L_SB: local salt-bridges; non_L: non-local salt bridges; II:_HSCI: intra- and interhelix, sheet and coil interactions; int_T: intervening residue table.
