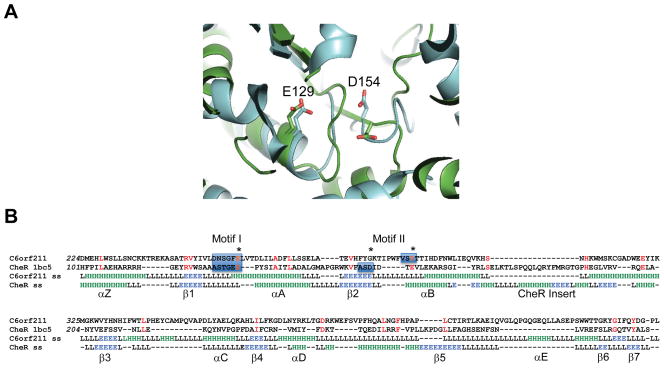
Figure 3. Structural similarities of the C6orf211 pocket with the SAM binding pocket of CheR.
(A) Structural superimpostions of S. cerevisiae protein YMR027W (3PT1.pdb) in cyan and S. typhimurium CheR (Uniprot code: P07801, PDB code: 1BC5.pdb) in green, revealing two acidic residues (E129 and D154 in CheR) in both proteins in similar positions within the active site. (B) Structure-based sequence alignment of human C6orf211 with S. typhimurium CheR. Conserved residues highlighted in red, stars indicate active site acidic residues, motifs I and II are highlighted with blue boxes. The first active site glutamate is conserved, the second, structurally equivalent acid residue occurs after a loop insert in C6orf211. I-Tasser predicted secondary structure shown for C6orf211 together with 1BC5.pdb secondary structure as defined by DSSP, green H indicates helix, blue E indicates strand and L is loop/coil. The conserved secondary structure elements in common with the core SAM-MT fold and the CheR insert are labeled. See also Figure S3.