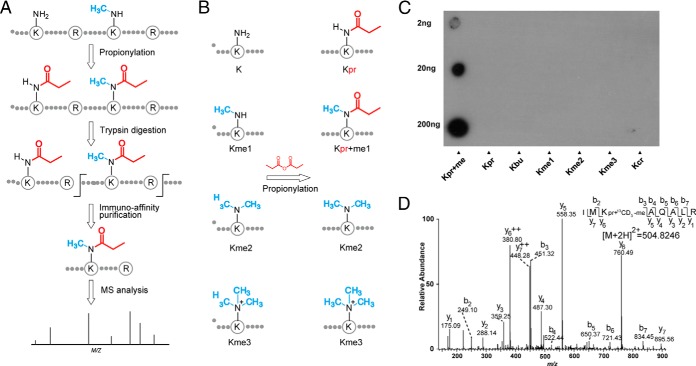
Fig. 1.
Strategy for monomethylome profiling. A, Schematic diagram of the experimental workflow for identifying monomethyllysine-containing peptides by chemical propionylation and immuno-affinity enrichment followed by MS analysis. B, The propionylation reaction occurs at monomethylated lysine but not di or trimethylated lysine. C, Specificity of the anti-propionyl monomethyllysineantibody by dot-blot assay. Kpr+me, Kpr, Kbu, Kme1, Kme2, Kme3, and Kcr denote peptide libraries of sequence xxxxxKxxxxx, where K represents a fixed lysine residue modified by propionyl-methyl, propionyl, butuyryl, monomethyl, dimethyl, trimethyl, and crotonyl groups, respectively. D, A typical annotated ion trap CID spectrum of a propionyl-13CD3-methyllysine-bearing peptide from protein HSP90.