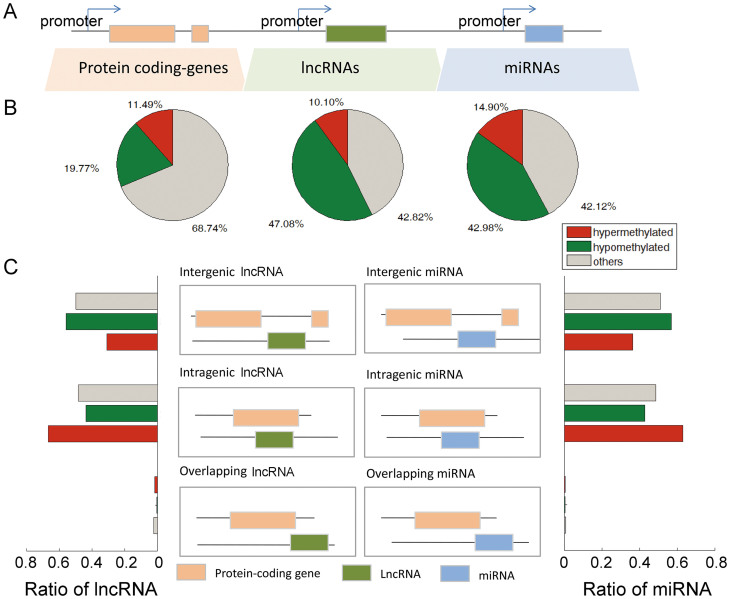
Figure 1. Characterization of the genome-wide methylation patterns of ncRNAs.
(A) Schematic representing the analyzed genomic features. (B) Pie charts representing the percentage of coding and non-coding promoters that overlapped with hyper- and hypo-methyalted DMRs in breast cancer. The green pie charts represent the percentage of the regopms covered by hypo-methylated DMRs, and the red pie charts represent the percentage of the regions covered by hypermethyalted DMRs. (C) LncRNAs and miRNAs were classified into different subcategories based on their location relative to protein-coding genes. The bar graphs show that the majority of intragenic ncRNAs were overrepresented with respect to hypermethylation and that most intergenic ncRNAs were hypomethylated in the breast cancer samples (all p-values < 0.05, Fisher's exact test).