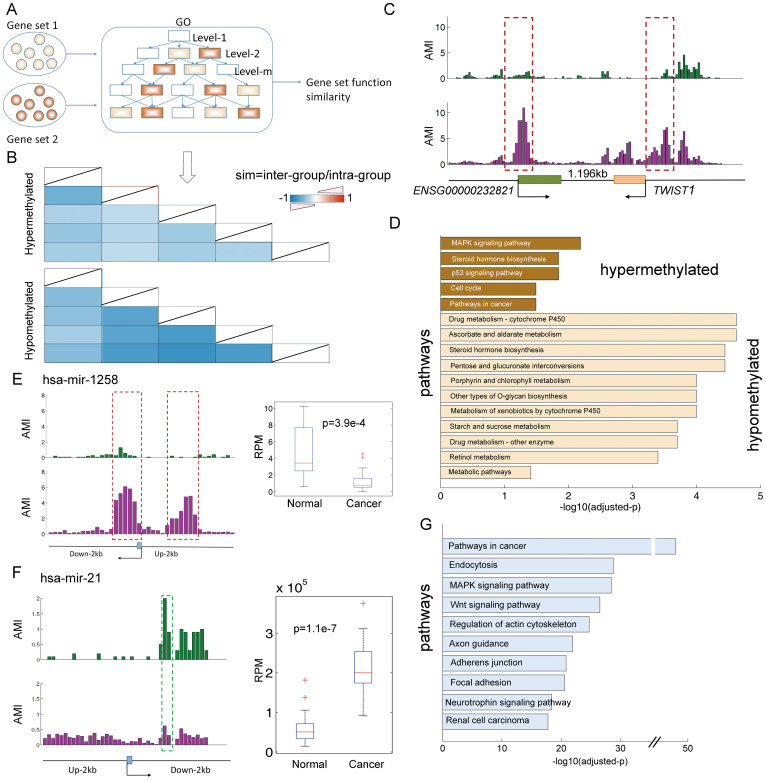
Figure 6. Aberrantly methylated ncRNAs widely disturb functions associated with breast cancer.
(A) The system used to calculate the gene set functional similarity. (B) The genes that displayed similar aberrant methylation patterns exhibited with high functional similarity. The similarity score was normalized to the intra-classes. (C) Representative lncRNA and protein-coding genes. The average methylation intensities (AMIs) in the control and breast cancer samples are shown. Green, control samples; Pink, cancer samples. (D) The biological processes enriched by adjacent genes of lncRNA displaying similar aberrant methylation patterns. (E) The average methylation and expression levels of hsa-mir-1258 in the normal and breast cancer samples. RPM, reads per million. (F) The average methylation and expression levels of hsa-mir-21 in the control and breast cancer samples. (G) The top 10 pathways enriched by targets of miRNA biomarkers.