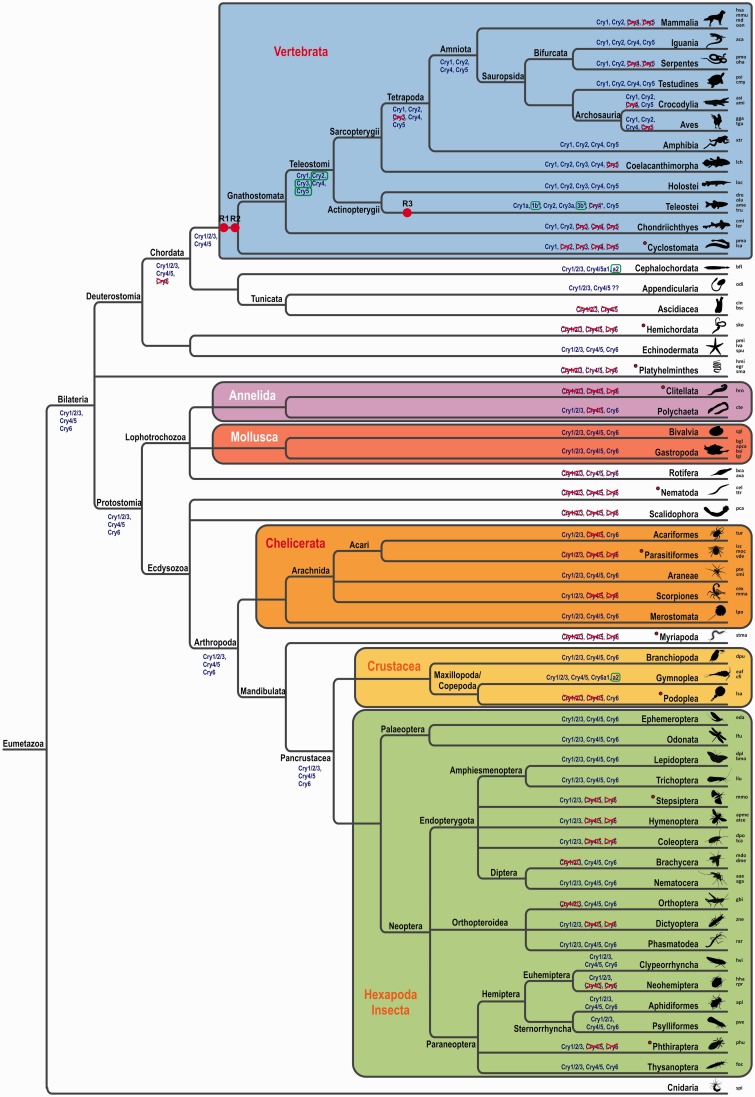
Fig. 2.—
Evolution of the cryptochrome genes. The phylogenetic relation of major subtypes of bilaterian species is represented in the depicted tree. Order, classes, and phylum names at branch points are indicated. Note that the branch lengths are NOT scaled with time. The major phyla and subphyla are highlighted by colored boxes. A representative image of one species for each order/class/phylum is given for better orientation. The abbreviation of the species used for phylogenetic reconstruction is shown at the right-hand side and corresponding species names are summarized in supplementary table S1, Supplementary Material online. Verified WGD events are indicated by red dots and labeled as R1, R2, and R3. Putative cryptochrome contents at extrapolated ancesteral stages are given at selected branch points and cryptochrome gains (green boxes) and losses (red crosses) are indicated. Note that parasitic or light independent species (red filled cycles) often lack cryptochrome genes or only have one variant left. Overall, multiple independent gene losses can be seen in isolated orders or classes.