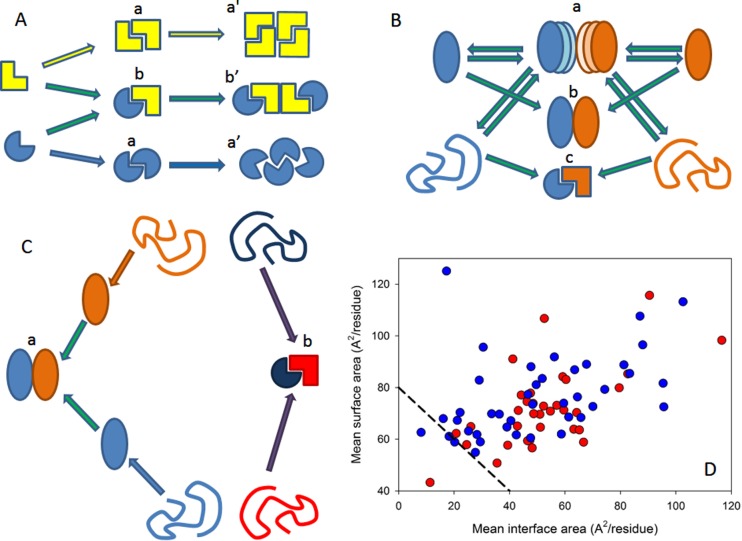
Figure 1.
Different classification types of protein–protein complexes. (A) Composition and geometry-based classifications. Complexes can be assembled from identical (a) and different subunits (b). Different types of monomers are shown by different shades of yellow and blue colors. Interactions leading to homo-oligomers are shown by arrows of the corresponding color. Interactions leading to the hetero-oligomers are shown by green arrows. Homodimers associate isologously. Interfaces of the dimers located at the center of homotetramers are also formed isologously, whereas all of the interfaces in the hetero-oligomers and the interfaces formed between the central homodimers and side-added monomers are formed heterologously. (B) Lifetime-based classification of oligomers. Complexes can be of transient (a), permanent nonobligate (b), or permanent obligate (c) nature. Formation of the permanent obligate complex is accompanied by the global folding of protomers. Hero-dimers and homologous transitions are shown for simplicity. (C) Folding-based classification. Protein complexes can be formed in a three-state mechanism (a), where protein folding and binding happen as two independent and subsequent steps. Alternatively, some proteins are formed in a two-state manner (b), where folding and binding occur simultaneously. (D) The per-residue surface area versus the per-residue interface area plot to discriminate between the three-state and two-state complexes. Here, the results of the computational disassembly of the eukaryotic ribosome (PDB ID: 3U5C and 3U5E)508 are shown. Surface and interface area normalized by the number of residues in each chain for the ribosomal proteins were estimated as described in ref (64). Proteins of the 40S and 60S subunits are shown by red and blue circles, respectively. A boundary separating ordered and disordered complexes is shown as a black dashed line.