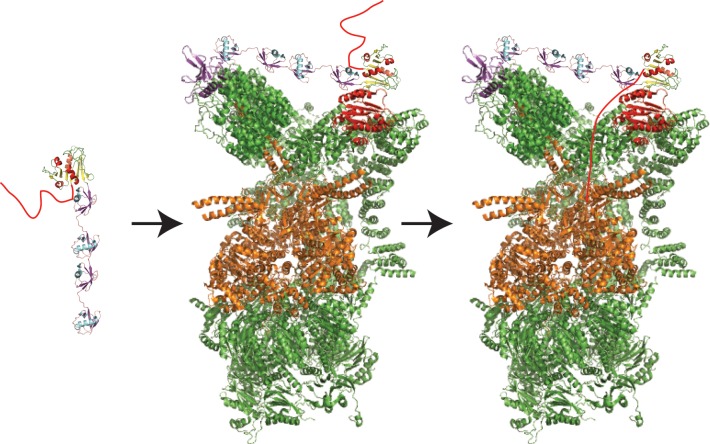
Figure 9.
Initiation of degradation by the proteasome requires a disordered region. A substrate molecule (dihydrofolate reductase, PDB ID 1DRE; yellow and red cartoon on the left) with a polyubiquitin chain attached (in this case, linear tetra-ubiquitin, from PDB ID 2W9N, purple and cyan cartoon on the left) and a disordered region (red tail) can be degraded by the proteasome. First the polyubiquitin modification docks at the proteasome (PDB ID 4C0V), presumably to ubiquitin receptors Rpn10 (red) and Rpn13 (purple), either simultaneously (as shown) or individually. Next, the tail is engaged by the Rpt ATPase motors (orange) in an ATP-dependent process, allowing unfolding, translocation, and degradation (along with deubiquitination of the substrate) to begin.