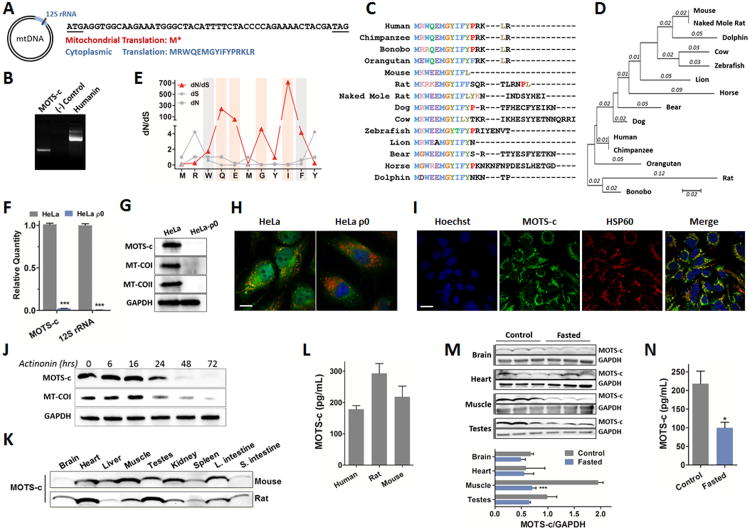
Figure 1. Identification of a novel expressed short open reading frame (sORF) encoded within the mitochondrial genome.
(A) MOTSC is encoded as a 51-bp short open reading frame (sORF) in the mitochondrial 12S rRNA. The mitochondrial genetic code yields tandem start/stop codons, whereas the cytoplasmic genetic code yields a viable peptide. (B) 3′ rapid amplification of cDNA ends (3′ RACE) of polyadenylated MOTS-c and humanin transcripts. (C-E) Phylogenetic analysis of MOTS-c with (C) multiple peptide sequence alignment in 14 species, and (D) its phylogenetic tree and branch length (estimated number of substitutions per site), and (E) the rate of non-synonymous (dN) and synonymous substitutions (dS), and their ratio (dN/dS) for the first well-conserved 11 residues of MOTS-c. The red bars represent residues that evolved under significant positive selection and the grey bars those under significant purifying selection. (F-H) HeLa-ρ0 cells are devoid of mitochondrial DNA. HeLa cells vs HeLa-ρ0 La cells: (F) 12S rRNA and MOTS-c transcripts by qRT-PCR, (G) MOTS-c peptide and mitochondrial-encoded cytochrome C oxidase I and II (MT-COI/II) protein, and nuclear-encoded GAPDH by immunoblotting, (H) MOTS-c (green), mitochondrial resident HSP60 (red), and nucleus (blue) by immunocytochemistry (ICC). Scale bar, 100-μm. (I) MOTS-c (green), HSP60 (red), and nucleus (blue) immunostaining in HEK293 cells. Scale bar, 20-μm. (J) Time-course detection of MOTS-c and MT-COI after treating with actinonin (150μM), which causes mitochondrial RNA degradation. (K-L) MOTS-c is detected in (K) various tissues in mice and rats and (L) circulation (plasma) in humans and rodents. (M-N) Fasting alters expression of endogenous MOTS-c in (M) tissues and (N) plasma in mice. Data shown as mean ± SEM. Student's t-test. *P<0.05, **P<0.01, ***P<0.001. See also Figure S1 and Table S1 for additional information on MOTS-c and validation for Western blotting, ICC, and ELISA.