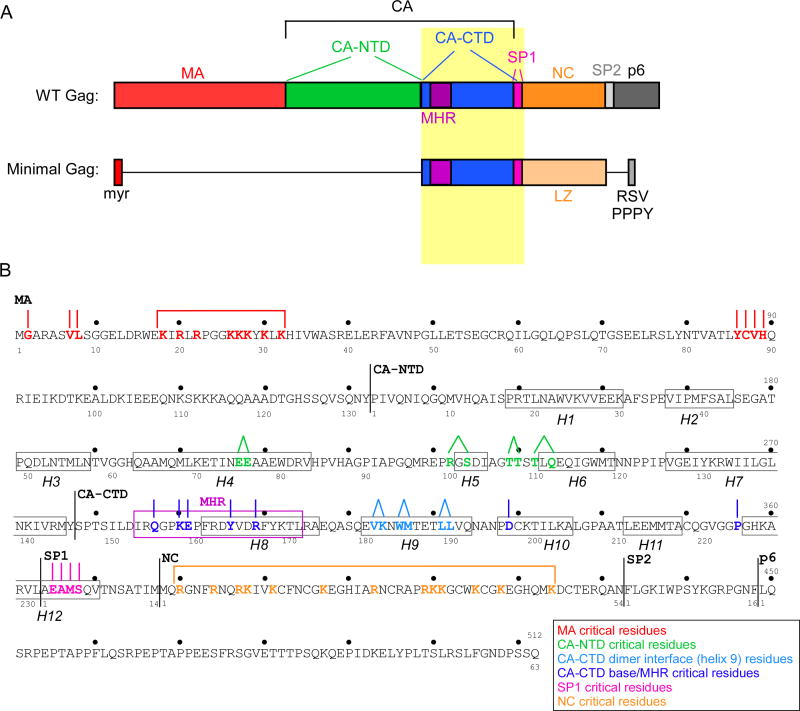
Figure 1. HIV-1 Gag domain map and amino acid sequence showing residues critical for immature capsid assembly.
(A) Schematic showing domains, subdomains, and spacer peptides in Gag [MA (red), CA-NTD (green), CA-CTD (blue), SP1 (pink), NC (orange), SP2 (light grey) and p6 (dark grey)]. The highly conserved MHR motif is highlighted in purple. Also shown is minimal Gag, a construct containing the minimal domains needed for assembly (Accola et al., 2000), in which some domains or subdomains are deleted (horizontal black lines), NC is replaced with a leucine zipper (LZ, light orange), and p6 is replaced with a short peptide containing the RSV PPPY motif (medium grey). B) Amino acid sequence of HIV-1 LAI Gag, with amino acid numbering for the complete Gag polyprotein shown above, and numbering for individual domains and spacer peptides (MA, CA, SP1, NC, and SP2) shown below. Note that the text and other figures in this review use the domain number system. Black lines indicate domain boundaries. The 12 actual or predicted α-helices in CA/SP1 are enclosed in black boxes (H1-H12). The MHR is enclosed in a purple box. Residues that are critical for immature capsid assembly are color-coded by surface (see legend). Lines above the critical residues indicate assembly-defective phenotypes produced by single (∣) or double (•) residue mutations; brackets indicate regions in which multiple basic residues are mutated to produce an assembly-defective phenotype.