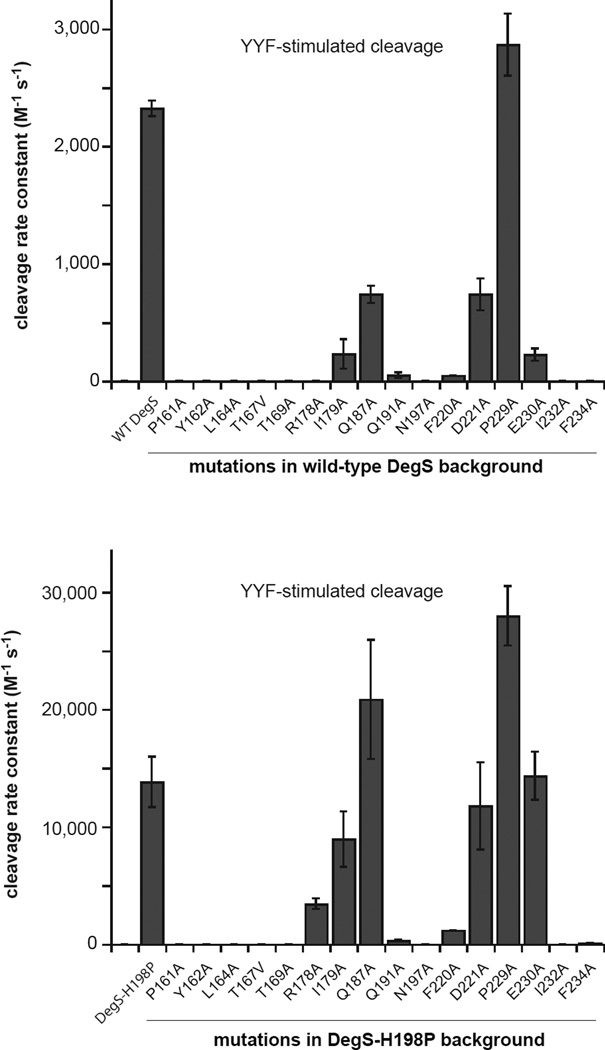
Figure 2. Mutational Effects on DegS Cleavage of RseA.
Effects of DegS mutations in an otherwise wild-type background (upper panel) or in an H198P background (lower panel) on OMP-peptide stimulated proteolytic activity, expressed as the second-order rate constant (kcat/KM) for RseA cleavage. Values are averages of two or more independent trials ± SEM. Cleavage reactions contained different sub-KM concentrations of 35S-labelled RseA, DegS or variants (1 µM trimer), and YYF OMP peptide (230 µM). Initial cleavage rates normalized by total enzyme were plotted as a function of the RseA concentration, and the second-order rate constant was determined from the slope of a linear fit.