Fig 1. Hic-5 was more abundant in immunoprecipitates of the Git1-Y554F mutant than in those of WT.
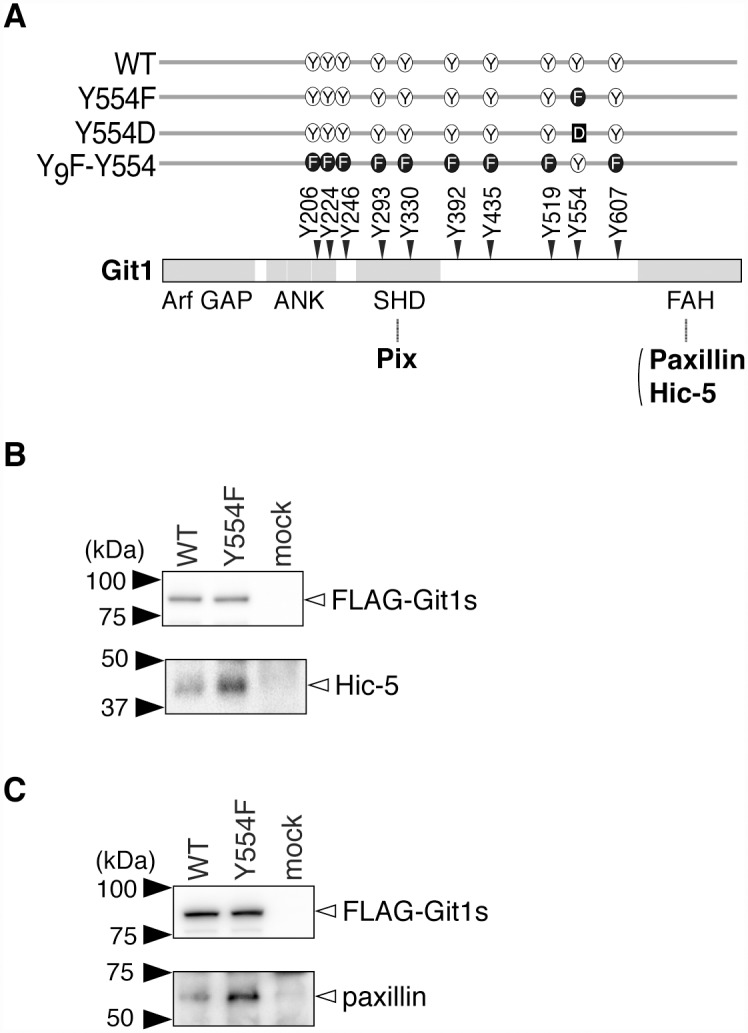
A, Schematic representation of Git1. The Git1 mutant constructs used in this study are shown with their abbreviated names. We previously showed that the replacement of ten potential phosphorylation tyrosine residues of Git1 with a phenylalanine residue resulted in the almost total disappearance of its tyrosine phosphorylation in cells, even after the pervanadate treatment, and then constructed Y9F-Y554 from the decuple mutant by changing Phe-554 back to tyrosine [12]. Pix binds to the SHD [4, 7–10], and Hic-5 and paxillin to the FAH [5, 6, 8]. Arf GAP, the GTPase-activating protein (GAP) domain for ADP-ribosylation factor (Arf); ANK, ankyrin repeats; SHD, Spa2 homology domain, FAH; focal adhesion targeting (FAT) homology domain. Y, tyrosine; F, phenylalanine; D, aspartic acid. B, C, Immunoprecipitation experiments. HEK293T cells expressing FLAG-tagged Git1 (WT), FLAG-tagged Git1-Y554F (Y554F), or a control vector (mock) were treated with 100 μM pervanadate for 15 min. The cell extracts were incubated with anti-FLAG beads, and the binding proteins were then specifically eluted with FLAG peptides. The eluates were separated by SDS-PAGE, followed by Western blotting with a rabbit anti-FLAG, mouse anti-Hic-5 (B), or anti-paxillin antibody (C).
