Figure 4.
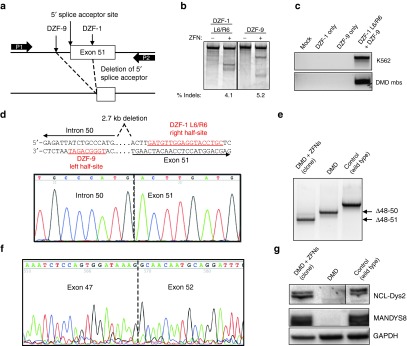
Restoration of the dystrophin reading frame in DMD patient myoblasts. (a) Schematic of strategy to delete exon 51 from the dystrophin gene locus. DZF-1 and DZF-9 flank the 5′ splice acceptor site of exon 51, which is removed after genomic deletion. P1/P2: primers used for detection of the genomic deletion by PCR in (c). (b) Gene modification activities of DZF-1 L6/R6 and DZF-9 as measured by the Surveyor assay 3 days after electroporation of 10 µg of each monomer expression cassette into DMD patient cells. (c) End-point genomic PCR across the deleted locus in human HEK293T or DMD myoblasts 3 days after treating cells with the indicated pair of nucleases. (d) Sanger sequencing of the PCR product from genomic DNA of a genetically corrected clonal cell population. Underlined sequences show target half-sites for the indicated ZFN target site. (e) End-point RT-PCR analysis of mRNA from control wild-type and untreated or a genetically corrected clonal population of DMD myoblasts after differentiation into myotubes. (f) Sanger sequencing of this PCR band showed the expected junction of exons 47 and 52. (g) Dystrophin expression as detected by western blot with antibodies to detect the C-terminus (NCL-DYS2) or rod domain (MANDYS8) in each of the indicated cell populations. All samples shown were run together on the same blot and cropped postimaging to remove extraneous lanes. However, different exposure times for the NCL-Dys2 western images were used to image DMD myoblasts and the genetically corrected clones compared to control samples to compensate for overexposure of control protein. The images for MANDYS8 and GAPDH are the same exposure time for all samples. DMD, Duchenne muscular dystrophy; ZFN, zinc finger nuclease.
