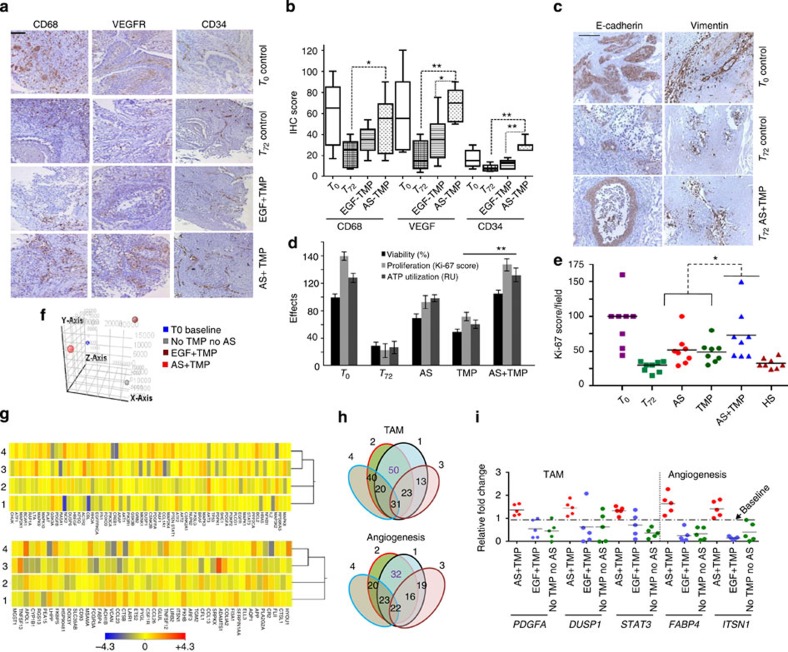
Figure 4. Integration of both TMP and AS in the CANScripts maintains the tumour ecosystem.
(a) Representative IHC images show the effect of AS and matchedTMP on the phenotypic stability of tumour explants 72 h post culture. Tumour sections were stained for CD68, VEGFR and CD34. Scale bar, 100 μm. (b) Quantitative IHC based box plot indicates CD68, VEGFR and CD34-positive cells in the explants maintained under different conditions. Horizontal line represents median and error bars indicate the interquartile range. *P<0.05 and **P<0.001, respectively (paired t-test, n=8). (c) Representative IHC images show EMT related markers of tumour microenvironment in the CANScript explants. Scale bar, 100 μm. (d) Graph shows the combined effects of AS and TMP on the functional integrity of the explants. Tumour sections were cultured for 72 h. Number of Ki-67-positive cells were counted and plotted along with percent viability and ATP utilization per section in triplicates (mean±s.d.). **P< 0.01 (by analysis of variance). (e) The combined effects of AS and TMP on the functional integrity of explants are represented as scatter plot (n=8). Number of Ki-67-positive cells were counted and plotted. HS was run as a control. *P< 0.05 (by paired t-test). (f) 3D-PCA plot showing global gene expression patterns between different culture conditions (that is, no AS and no TMP, EGF+TMP, AS+TMP and T0 baseline) obtained from HNSCC tumour explants after 12 h. After initial normalization of data analysis was performed compared with baseline. (g) Heat map analysis of the microarray data showing the genes related to TAM (top) and angiogenesis (bottom). Tumours explants were cultured in TMP-coated plates with AS (AS+TMP, lane 2) or EGF (EGF+TMP, lane 3) or in uncoated plates without AS (No TMP and no AS Control, lane 4) and transcriptomic pattern was compared with base line tumour (lane 1). Heat map scale indicates the expression range. Clustering of genes was performed by k- means algorithm. Distance was measured by Euclidean distance metric. (h) Venn diagram showing number of overlapped genes related to TAM and angiogenesis between the three culture conditions. (i) Validation of microarray gene signature by qRT–PCR for TAM (left) and angiogenesis (right); selected genes from each signature was run in triplicates (technical replicates) normalized to baseline expression (biological replicates) and compared between conditions as indicated in the scatter plot (n=5).