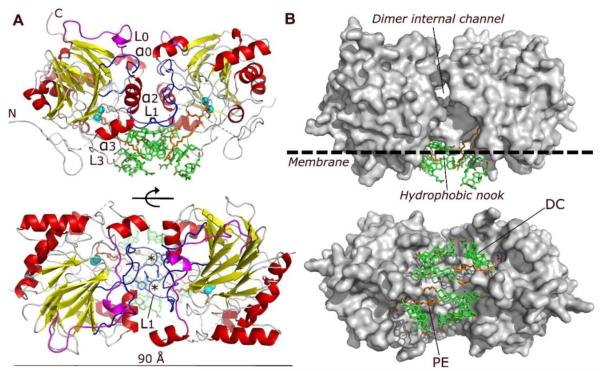
Figure 1. Structure of human NAPE-PLD.
(A) Ribbon trace of the NAPE-PLD dimer, front view (top) and bottom view (bottom). Dimer subunits interact mainly through their loops L0 (magenta) and L1 (blue), structural elements that distinguishes NAPE-PLD from other MβL proteins. The subunit orientation produces a symmetric alignment of loops L1, which extend in the dimer the existing monomer binding faces. The active sites contain a binuclear zinc metal center (spheres, color cyan), which faces the lipid bilayer and binds a molecule of PE (carbon atoms in orange). PE derives from the expression system used to produce NAPE-PLD and occupies the binding site reserved to the PE fragment of NAPE. Several molecules of the non-conjugated C24 bile acid DC (carbon atoms in green) bind to the enzyme, creating a lipid substructure at the interface with the membrane. N and C indicate the N-terminus and C-terminus of the protein, respectively. The asterisks indicate the position of L1 residues Q158 and Y159. (B) Surface representation of the NAPE-PLD dimer shown with the same orientation as in (A). An internal channel with a diameter of ~9 Å partly separates the two subunits of NAPE-PLD, and overlooks a hydrophobic nook to which PE and DC are bound. The predicted membrane boundary is shown as dotted line.