Figure 4.
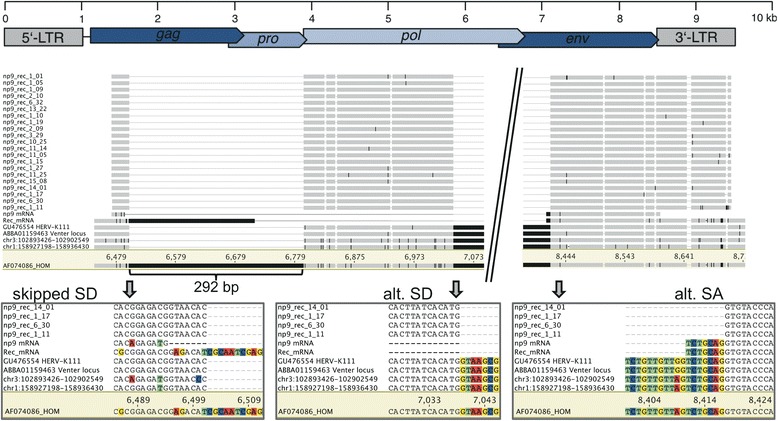
Excerpts from a multiple sequence alignment of an alternative exon from one or several HERV-K(HML-2) type 1 loci. The utilized SD site for the np9 intron 2 (‘alt. SD’) is located more downstream compared to the canonical np9 SD2 (‘skipped SD’). The 3′ end of intron 2 is also located 7 nt more downstream compared to the canonical np9/rec mRNA SA2 (‘alt. SA’). Several sequences have been included for comparison: canonically spliced ‘np9_mRNA’ and ‘Rec_mRNA’ [7,8], corresponding sequence portions from the HERV-K111 provirus [32], the Venter locus (GenBank acc. no. ABBA01159463; ref. [33]), and the HERV-K(HML-2.HOM) (type 2) provirus (GenBank acc. no. AF074086; ref. [31]) (nt positions of that sequence are given as reference), HML-2 loci ERVK-5 and ERVK-18 in human chromosomes 3 and 1 (nt positions according to hg18) as most closely related, yet, clearly different HML-2 proviruses (see the paper text). Most of the np9/rec intron 2 has been omitted from the figure, as indicated. Nucleotide positions deviating from the consensus sequence of included sequences are highlighted in black. Enlarged alignment regions shown at the bottom depict usage of a different SD2 site compared to np9 mRNA (left and center) and usage of a different SA2 compared to rec and np9 mRNA. Note the mutated SA2 site (AG → GG) in the HERV-K111 and Venter locus sequences (see the paper text). A HERV-K(HML-2) provirus map with locations of ORFs is shown at the top. LTR, long terminal repeat; SA, splice acceptor; SD, splice donor.
