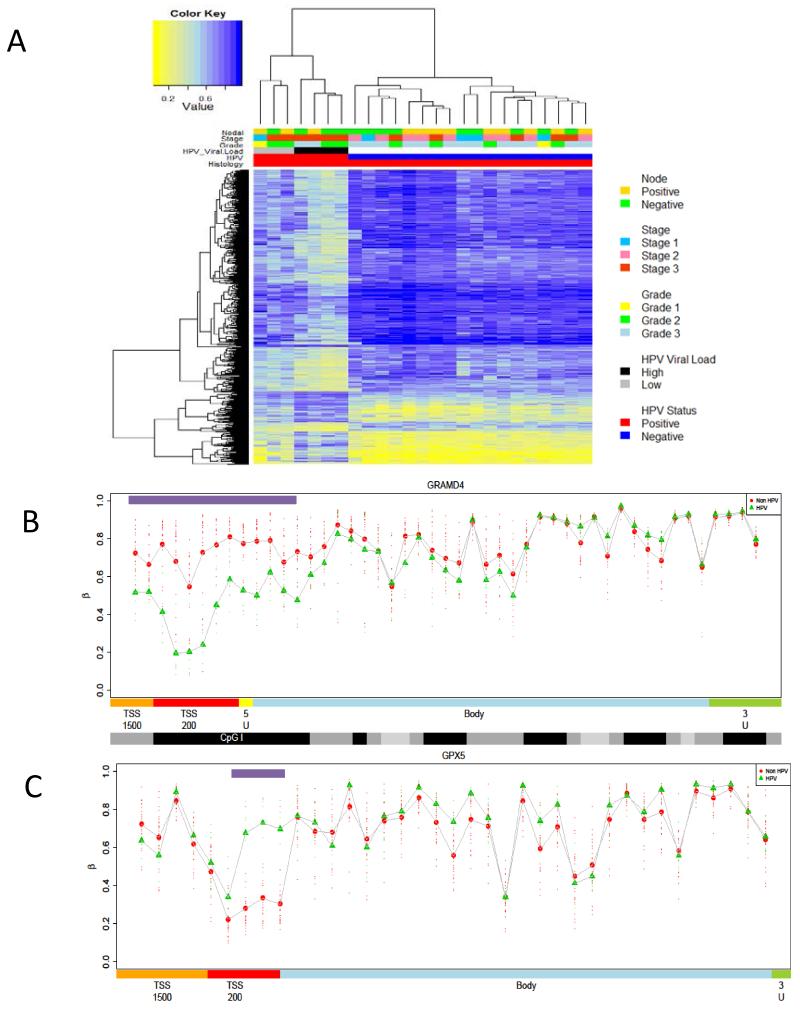
Figure 5. PeCa HPV induced epigenetic signature.
A) Heat map of significant MVP (p<0.01) between HPV positive and HPV negative PeCa. HPV positive samples show a significantly lower methylation profile than HPV negative disease. B,C) Methylation profiles of candidate genes epigenetically deregulated during HPV tumorigenic transformation. Comparison of DMR profiles across canonical features for HPV associated PeCa (green) and non-HPV associated PeCa (red), for candidate epigenetically regulated genes involved in the HPV driven penile cancer ((B)GPX5 and (C)GRAMD4). Profiles show Feature annotation is as provided by BeadChip, and methylation values are colour-coded accordingly: orange = TSS1500, (1500 bp to 200 bp upstream of the transcription start site (TSS)); red = TSS200 (200 bp upstream of the TSS,); yellow = 5′ untranslated region (UTR), blue = gene body; black = CpG islands,; darker grey = CpG shores,; and light grey = CpG shelves