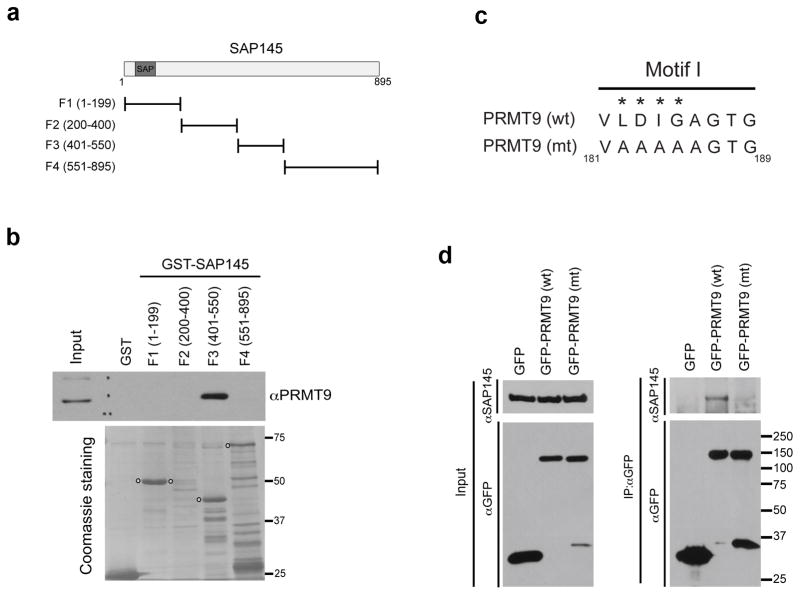
Fig. 3. Mapping the interaction regions of PRMT9 and SAP145.
(a) A series of GST-fusion truncations of SAP145 were generated. The location of the SAP (SAF-A/B, Acinus and PIAS) domain is indicated.
(b) PRMT9 interacts with the amino acids 401–550 of SAP145. GST pull-down experiment was performed by incubating HeLa cell total lysates with purified GST or GST-tag SAP145 fragments described in (A). The pull-down samples were eluted and detected by western blotting using αPRMT9 antibody. The loading of the proteins was visualized by coomassie staining of the membrane. The open circles indicate the individual GST-tag SAP145 fragments.
(c) Generation of enzymatic mutant PRMT9. The amino acids (LDIG) located within the conserved Motif I of PRMT9 were mutated to AAAA, causing the loss of methyltransferase activity of enzyme. The numbers below indicate the location of the amino acids on the protein.
(d) Wild type, but not the enzymatic mutant, PRMT9 interacts with SAP145. Co-immunoprecipitation was performed in HeLa cells transiently transfected with GFP control vector, GFP-PRMT9 (wt) and GFP-PRMT9 (mt), as described in (c). Total cell lysates were immunoprecipitated with αGFP antibody. The eluted protein samples were detected by western blotting with αSAP145 and αGFP antibodies. The input samples were detected with αSAP145 and αGFP antibodies as well.