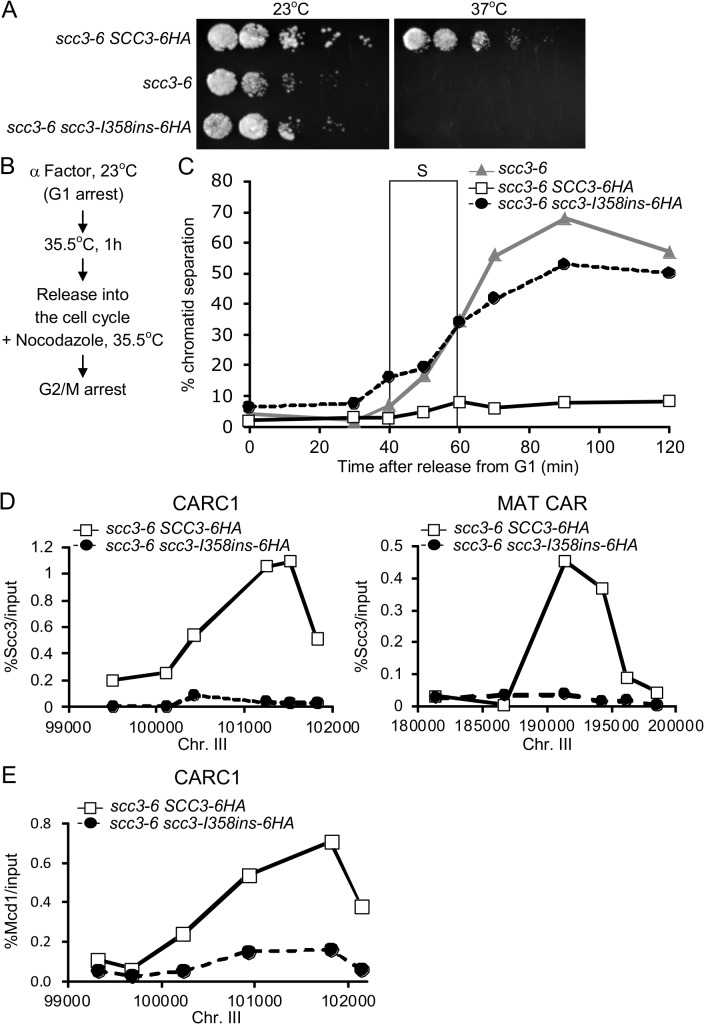
Fig 3. Cohesion and chromosome binding of scc3-I358Ins under native expression levels.
A. Strains YIO81 (scc3-6), YIO91 (SCC3-6HA scc3-6) andYIO91R1 (scc3-I358ins-6HA scc3-6) were grown to saturation in YPD media. Fivefold serial dilutions of each strain was plated on YEPD plates and grown at either the permissive (23°C) or restrictive (37°C) temperature for scc3-6. B. Flowchart of the experimental design to score sister chromatid cohesion. C. Strains as indicated in A were grown at 23°C to mid-log phase and arrested in G1 using α-factor. Cells were then shifted to 35.5°C for 1 h, released into the cell cycle and re-arrested in G2/M with nocodazole. Samples for the cohesion assay were taken approximately every 15 minutes (n = 3). The frame indicates S-phase of the cell cycle as determined by flow cytometry (S1 Fig). D. Strains described in A were processed as in B for chromatin immunoprecipitation analysis. Scc3, HA tagged proteins were immunoprecipitated. Precipitated DNA was analyzed by quantitative PCR with six primer pairs for the MAT CAR and CARC1, as described (Material and methods). A representative experiment is shown (n = 3). E. Strains described in A were processed as in B for chromatin immunoprecipitation analysis. Mcd1 was immunoprecipitated. Precipitated DNA was analyzed by quantitative PCR for CARC1. A representative experiment is shown (n = 3).