Figure 2.
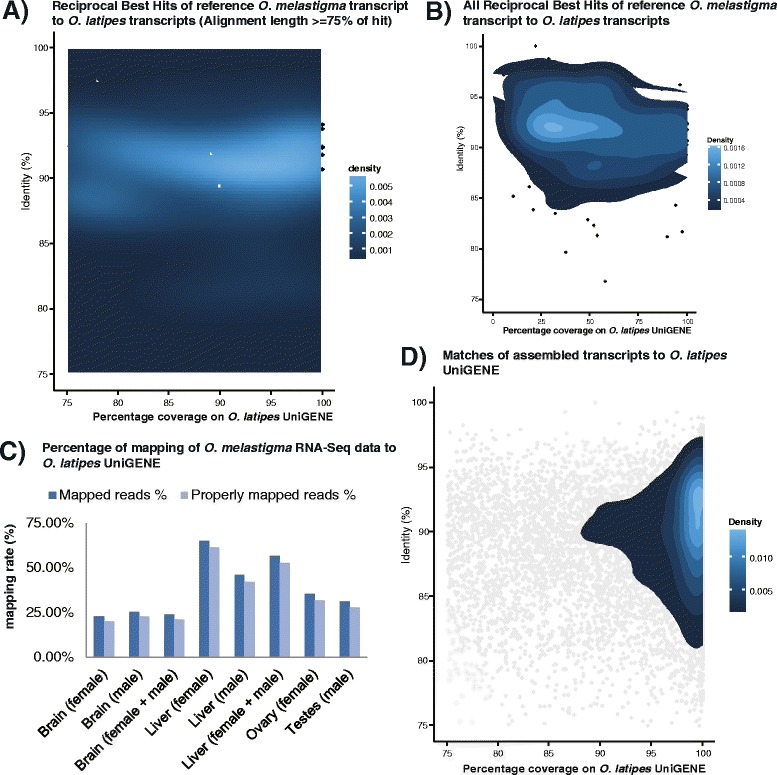
Transcriptome divergence between marine and freshwater medaka. A) Nucleotide identity between Oryzias melastigma and Oryzias latipes orthologs deposited in the NCBI nucleotide database. Only those with an alignment length >75% are shown. B) Nucleotide identity between all O. melastigma and O. latipes orthologs. C) Low mapping rate of O. melastigma RNA-Seq data generated in this study onto the O. latipes UniGENE dataset. The mapping rate is highest for liver and lowest for brain. D) The nucleotide identity between transcripts (Ensemble Assembly) assembled in this study and the O. latipes UniGENE dataset. Only those with alignment length >75% are shown.
